On a Sunday morning when you want to learn, try adding a hard disk to the KVM virtual machine with the command line.
create a disk
#qemu-img create – F qcow2/home/KVM FS/sy-b80915disk1.qcow2 10g
Bind disk to domain: sy-b80915
#virsh attach disk sy-b80915/home/KVM FS/sy-b80915disk1.qcow2 VDB — live — config
Later, I tried to unbind the VDA of the main disk. As a result, I accidentally unbind the VDA of the main disk
#virsh detach disk sy-b80915 VDA — live — config
But I unbound VDB, namely sy-b80915disk1.qcow2
#virsh detach disk sy-b80915 VDB — live — config
The virtual machine can still be restarted and used normally later, but I think the name of sy-b80915disk1.qcow2 is not good. Delete and recreate sy-b80915vdb.qcow2
#rm – RF/home/KVM/sy-b80915disk1.qcow2
#qemu-img create – F qcow2/home/KVM FS/sy-b80915vdb.qcow2 10g
Then bind
#virsh attach disk sy-b80915/home/KVM FS/sy-b80915vdb.qcow2 VDB — live — config
the results are as follows:
error: XML error: target ‘VDB’ duplicated for disk sources’ sy-b80915disk1. Img ‘and’ sy-b80915vdb. Img ‘
the big idea is to bind repeatedly, but it has been unbound before.
The only exception is that the main disk VDA is unbound accidentally, but the system can still run. So I checked the XML file. Compared with other virtual machines, I found that the XML file of SY – b80915 lacks the main disk VDA. When unbinding the VDA, the XML file is changed. So add VDA to the XML file again
execute the following command to edit the XML file:
#virsh edit sy-b80915
repair the XML definition of VDA, as shown in Figure 1:
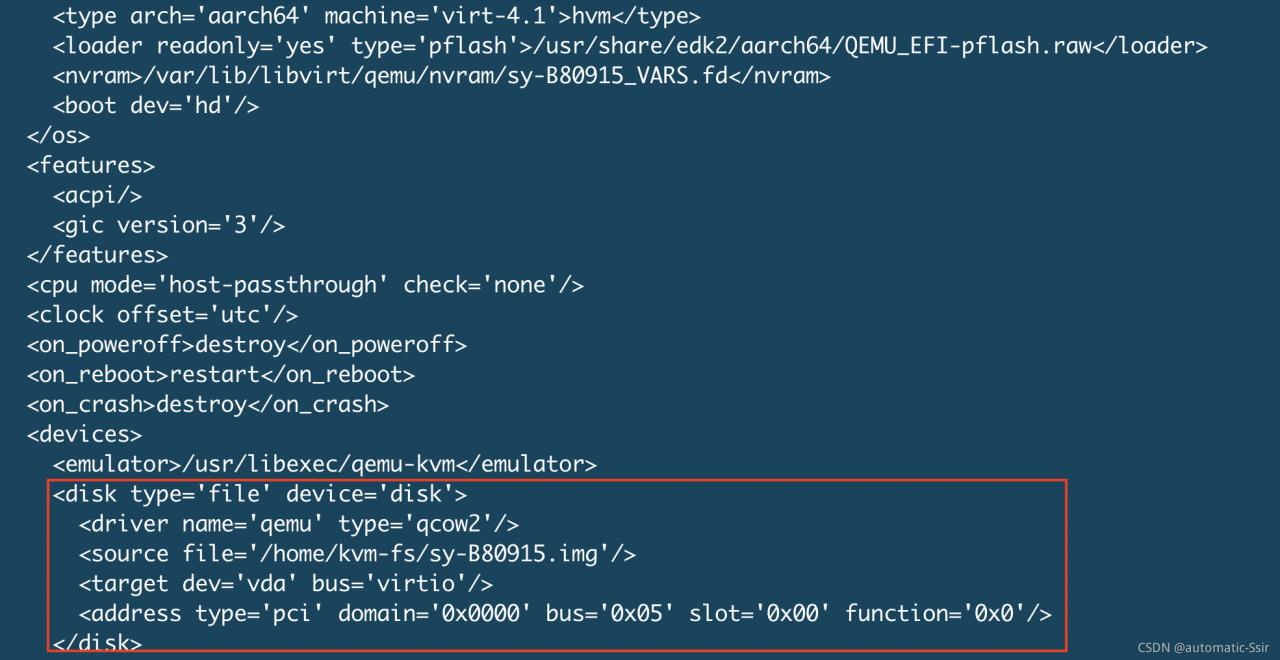
Figure 1
Then bind sy-b80915vdb.qcow2 again. Success
#virsh attach-disk sy-B80915 /home/kvm-fs/sy-B80915vdb.qcow2 vdb –live –config