This is because the R language default behavior is a unique identifier
Duplicate lines need to be removed
This is very common in differential expression, because different IDs may correspond to the same gene_ symbol
genes_sig <- res_sig %>%
arrange(adj.P.Val) %>% #Sort by Pvalue from smallest to largest
as tibble() %>%
column_to_rownames(var = "gene_symbol")report errors
Error in `.rowNamesDF<-`(x, value = value) :
duplicate 'row.names' are not allowed
In addition: Warning message:
non-unique values when setting 'row.names': ‘’, ‘ AMY2A ’, ‘ ANKRD20A3 ’, ‘ ANXA8 ’, ‘ AQP12B ’, ‘ AREG ’, ‘ ARHGDIG ’, ‘ CLIC1 ’, ‘ CTRB2 ’, ‘ DPCR1 ’, ‘ FAM72B ’, ‘ FCGR3A ’, ‘ FER1L4 ’, ‘ HBA2 ’, ‘ HIST1H4I ’, ‘ HIST2H2AA4 ’, ‘ KRT17P2 ’, ‘ KRT6A ’, ‘ LOC101059935 ’, ‘ MRC1 ’, ‘ MT-TD ’, ‘ MT-TV ’, ‘ NPR3 ’, ‘ NRP2 ’, ‘ PGA3 ’, ‘ PRSS2 ’, ‘ REEP3 ’, ‘ RNU6-776P ’, ‘ SFTA2 ’, ‘ SLC44A4 ’, ‘ SNORD116-3 ’, ‘ SNORD116-5 ’, ‘ SORBS2 ’, ‘ TNXB ’, ‘ TRIM31 ’, ‘ UGT2B15 ’ Try to use the duplicated() function
res_df = res_df[!duplicated(res_df),] %>% as.tibble() %>%
column_to_rownames(var = "gene_symbol") Still not?Duplicated removing duplicate values still shows that duplicate values exist
Error in `.rowNamesDF<-`(x, value = value) :
duplicate 'row.names' are not allowed
In addition: Warning message:
non-unique values when setting 'row.names': ‘ AMY2A ’, ‘ ANKRD20A3 ’, ‘ ANXA8 ’, ‘ AQP12B ’, ‘ AREG ’, ‘ ARHGDIG ’, ‘ CLIC1 ’, ‘ CTRB2 ’, ‘ DPCR1 ’, ‘ FAM72B ’, ‘ FCGR3A ’, ‘ FER1L4 ’, ‘ HIST1H4I ’, ‘ HIST2H2AA4 ’, ‘ KRT17P2 ’, ‘ KRT6A ’, ‘ LOC101059935 ’, ‘ MT-TD ’, ‘ MT-TV ’, ‘ NPR3 ’, ‘ NRP2 ’, ‘ PGA3 ’, ‘ PRSS2 ’, ‘ REEP3 ’, ‘ RNU6-776P ’, ‘ SFTA2 ’, ‘ SORBS2 ’, ‘ TRIM31 ’, ‘ UGT2B15 ’ Another way is to use the uniqe function of the dyplr package
res_df = res_df %>% distinct(gene_symbol,.keep_all = T) %>% as.tibble() %>%
column_to_rownames(var = "gene_symbol") Successfully solved

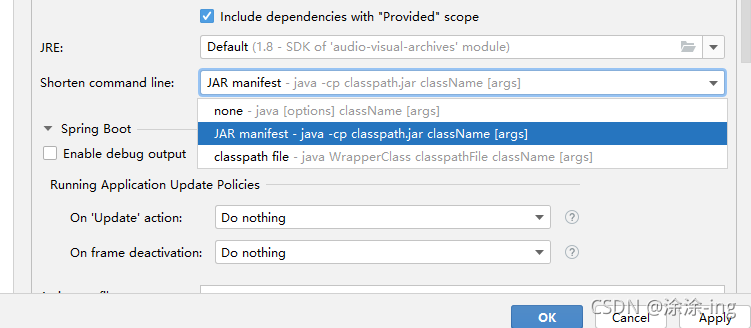
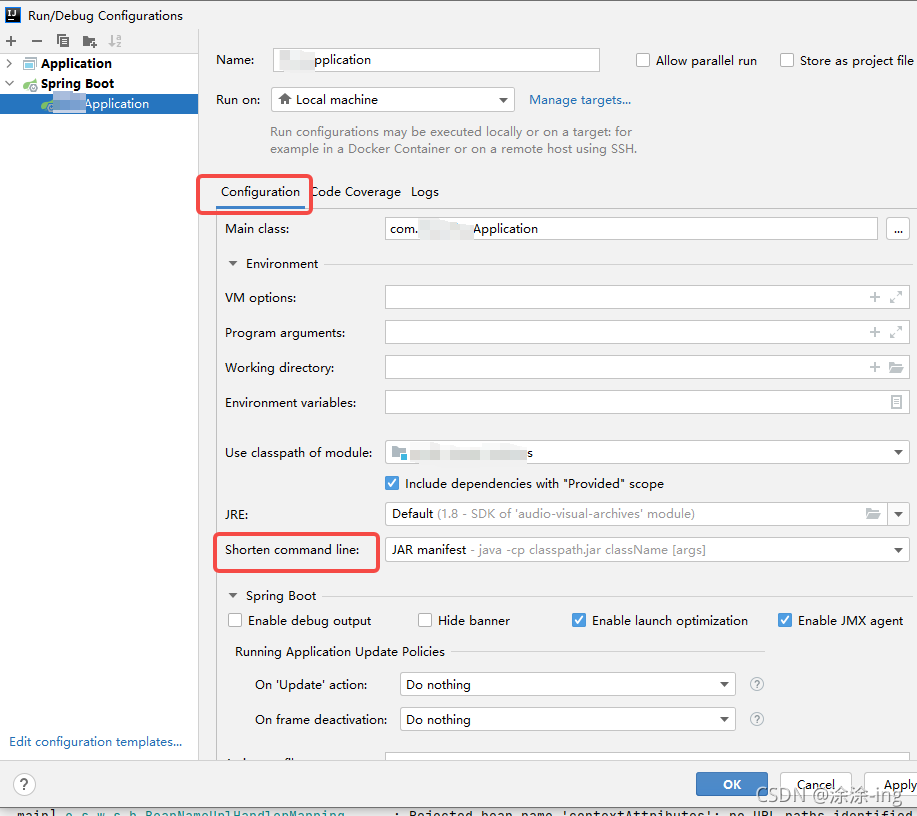 in the short command line option, select the jar manifest option, because the default is the first none,
in the short command line option, select the jar manifest option, because the default is the first none,