Question:
##Boundary fill
import cv2
top_size,bottom_size,left_size,right_size=(50,50,50,50)
replicate=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_REPLICATE)
reflect=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_REFLECT)
reflect101=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_REFLECT101)
wrap=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_WRAP)
constant=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_CONSTANT,value=0)
Outcome:
error Traceback (most recent call last)
<ipython-input-39-820a457b4770> in <module>
2 import cv2
3 top_size,bottom_size,left_size,right_size=(50,50,50,50)
----> 4 replicate=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_REPLICATE)
5 reflect=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_REFLECT)
6 reflect101=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,boderType=cv2.BORDER_REFLECT101)
error: OpenCV(4.5.2) :-1: error: (-5:Bad argument) in function 'copyMakeBorder'
> Overload resolution failed:
> - copyMakeBorder() missing required argument 'borderType' (pos 6)
> - copyMakeBorder() missing required argument 'borderType' (pos 6)
analysis:
error: OpenCV(4.5.2) :-1: error: (-5:Bad argument) in function 'copyMakeBorder'
> Overload resolution failed:
> - copyMakeBorder() missing required argument 'borderType' (pos 6)
> - copyMakeBorder() missing required argument 'borderType' (pos 6)
“Bordertype” spelling error!!!!
Correct code:
##Boundary fill
import cv2
top_size,bottom_size,left_size,right_size=(50,50,50,50)
replicate=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,borderType=cv2.BORDER_REPLICATE)
reflect=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,borderType=cv2.BORDER_REFLECT)
reflect101=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,borderType=cv2.BORDER_REFLECT101)
wrap=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,borderType=cv2.BORDER_WRAP)
constant=cv2.copyMakeBorder(img,top_size,bottom_size,left_size,right_size,borderType=cv2.BORDER_CONSTANT,value=0)
import matplotlib.pyplot as plt
plt.subplot(231),plt.imshow(img,'gray'),plt.title('ORIGINAL')
plt.subplot(232),plt.imshow(replicate,'gray'),plt.title('REPLICATE')
plt.subplot(233),plt.imshow(reflect,'gray'),plt.title('REFLECT')
plt.subplot(234),plt.imshow(reflect101,'gray'),plt.title('REFLECT101')
plt.subplot(235),plt.imshow(wrap,'gray'),plt.title('WRAP')
plt.subplot(236),plt.imshow(constant,'gray'),plt.title('CONSTANTL')
plt.show()
Output results:
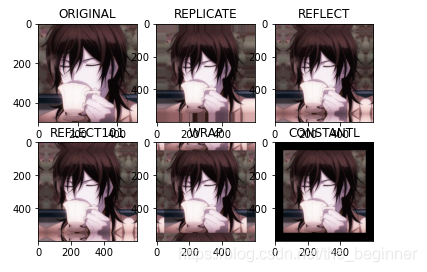
cv2.BORDER_ Replicate copy method, copy the edge pixel CV2. Border_ Reflect reflection method, the interested image pixels on both sides of the entry CV2. Border_ Reflect101 reflection method, with the most edge pixel as the axis, symmetrical CV2. Border_Wrap method cv2.border_Constant good method