Solution:
FileNotFoundError: Could not find module
‘D:\Anaconda3\envs\LabelImg\label\bin\geos_c.dll’ (or one of its dependencies).
Try using the full path with constructor syntax.(or one of its dependencies). Try using the full path with constructor syntax.
1, most people are not importing the “shapely” dependency. You only need to import it
pip install shapely
2. Import this or report an error, basically it is wrong with your Python version.
To download the corresponding. WHL file, the link is as follows: https://www.lfd.uci.edu/~gohlke/pythonlibs/#shapely , after entering the website, Ctrl + F can find shapely, and select the version up or down according to your own version( The author is python3.8)
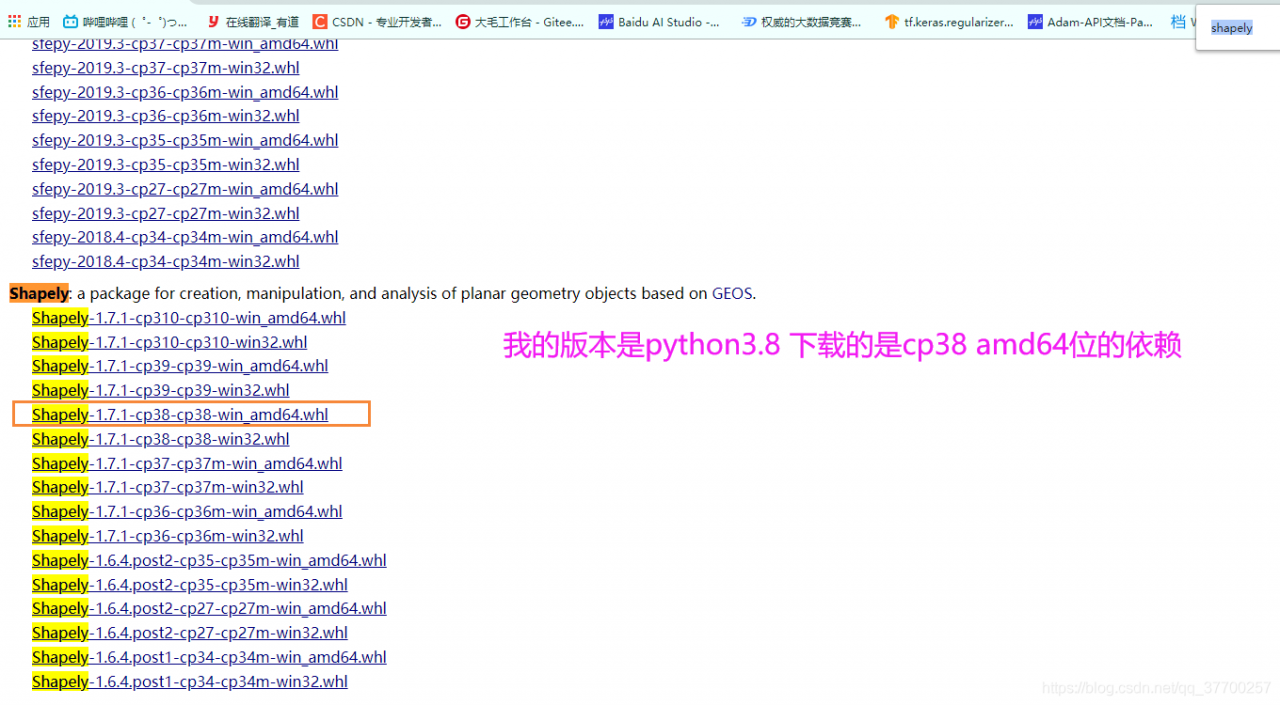
#First uninstall the downloaded shapely
pip uninstall shapely
#then download the local cp38 version of shapely
pip install D:\PyCharm\Pro\OCR\PaddleOCR\PPOCRLabel\BACK\Shapely-1.7.1-cp38-cp38-win_amd64.whl
 tsinghua source: https://pypi.tuna.tsinghua.edu.cn/simple/opencv-python/
tsinghua source: https://pypi.tuna.tsinghua.edu.cn/simple/opencv-python/