When debugging the code of densenet for classification task, the following errors are encountered in the process of image preprocessing:
runtimeerror: stack expectations each tensor to be equal size, but got [640, 640] at entry 0 and [560, 560] at entry 2
it means that the size of the loaded sheets is inconsistent
after searching, it is found that there should be a problem in the preprocessing process when I load the image
the following is the instantiation part of training data preprocessing.
train_transform = Compose(
[
LoadImaged(keys=keys),
AddChanneld(keys=keys),
CropForegroundd(keys=keys[:-1], source_key="tumor"),
ScaleIntensityd(keys=keys[:-1]),
# # Orientationd(keys=keys[:-1], axcodes="RAI"),
Resized(keys=keys[:-1], spatial_size=(64, 64), mode='bilinear'),
ConcatItemsd(keys=keys[:-1], name="image"),
RandGaussianNoised(keys=["image"], std=0.01, prob=0.15),
RandFlipd(keys=["image"], prob=0.5), # , spatial_axis=[0, 1]
RandAffined(keys=["image"], mode='bilinear', prob=1.0, spatial_size=[64, 64], # The 3 here is because we don't know what the size of the three modal images will be after stitching, so we first use
rotate_range=(0, 0, np.pi/15), scale_range=(0.1, 0.1)),
ToTensord(keys=keys),
]
)
My keys are [“t2_img”, “dwi_img”, “adc_img”, “tumor”]
the error shows that the loaded tensor has dimensions [640, 640] and [560, 560], which are the dimensions of my original image, which indicates that there may be a problem in my clipping step or resize step. Finally, after screening, it is found that there is a problem in my resize step. In the resize step, I selected keys = keys [: – 1], that is, it does not contain “tumor”. Therefore, when resizing, my tumor image will still maintain the size of the original image, and the data contained in this dictionary will still be a whole when loaded, The dimensions of each dimension of the whole will automatically expand to the largest of the corresponding dimensions of all objects, so the data I loaded will still be the size of the original drawing. Make the following corrections:
train_transform = Compose(
[
LoadImaged(keys=keys),
AddChanneld(keys=keys),
CropForegroundd(keys=keys[:-1], source_key="tumor"),
ScaleIntensityd(keys=keys[:-1]),
# # Orientationd(keys=keys[:-1], axcodes="RAI"),
Resized(keys=keys, spatial_size=(64, 64), mode='bilinear'), # remove [:-1]
ConcatItemsd(keys=keys[:-1], name="image"),
RandGaussianNoised(keys=["image"], std=0.01, prob=0.15),
RandFlipd(keys=["image"], prob=0.5), # , spatial_axis=[0, 1]
RandAffined(keys=["image"], mode='bilinear', prob=1.0, spatial_size=[64, 64], # The 3 here is because we don't know what the size of the three modal images will be after stitching, so we first use
rotate_range=(0, 0, np.pi/15), scale_range=(0.1, 0.1)),
ToTensord(keys=keys),
]
)
Run successfully!


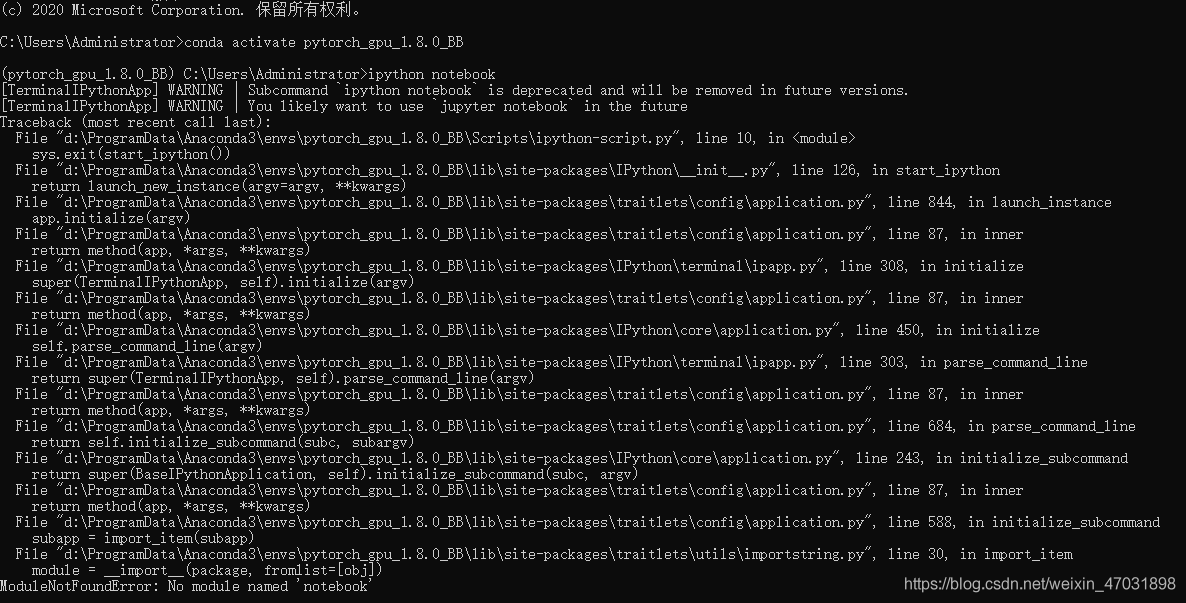
 activate the environment when you run the code: “CONDA activate + your environment name”
activate the environment when you run the code: “CONDA activate + your environment name” 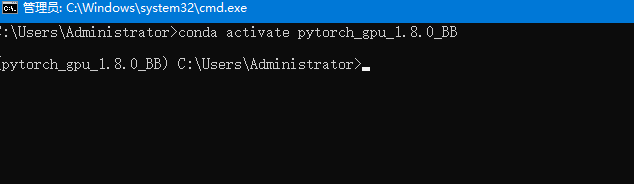 after entering your environment, enter “Python – M PIP install Jupiter”, and then “enter”
after entering your environment, enter “Python – M PIP install Jupiter”, and then “enter”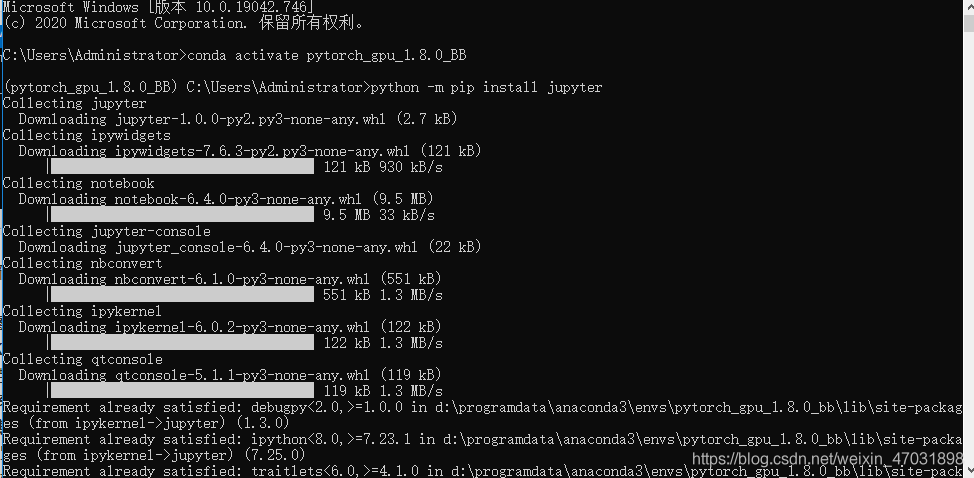 appears at the bottom to indicate that the installation is successful
appears at the bottom to indicate that the installation is successful 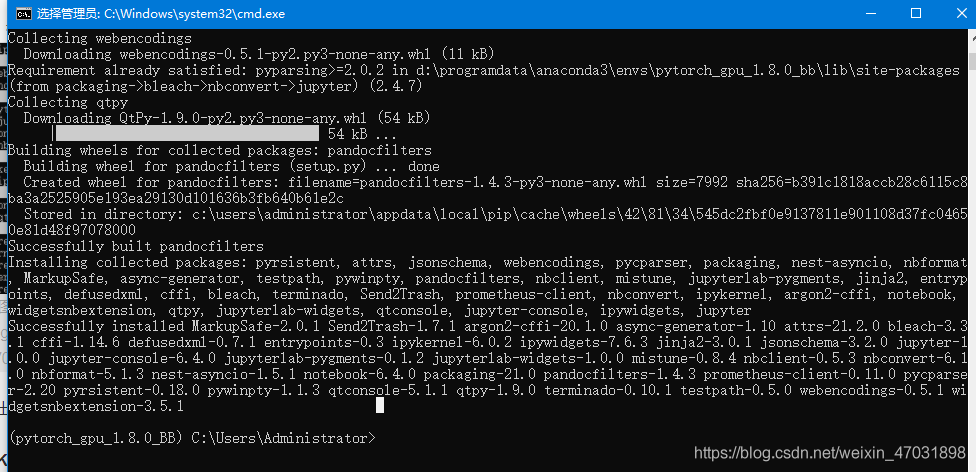 then enter: IPython notebook
then enter: IPython notebook 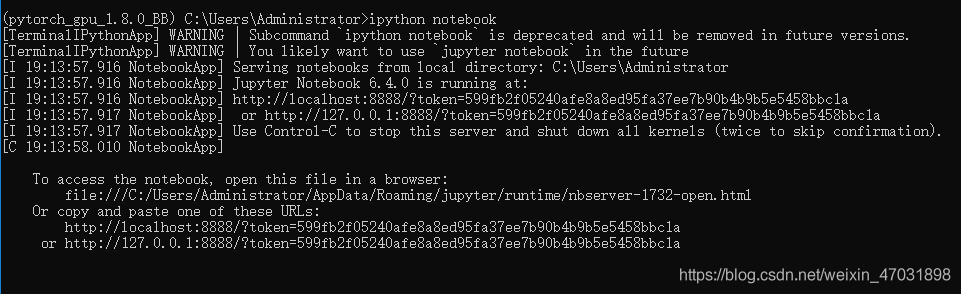 . This page indicates that the problem has been solved
. This page indicates that the problem has been solved