After I read the data from the mat file with python, I get a dictionary array. I want to store this dictionary array in a json file, so the json data should be encoded first, so the json.dumps function is used for encoding, but I use json. It is found that there will be problems during the dumps function:
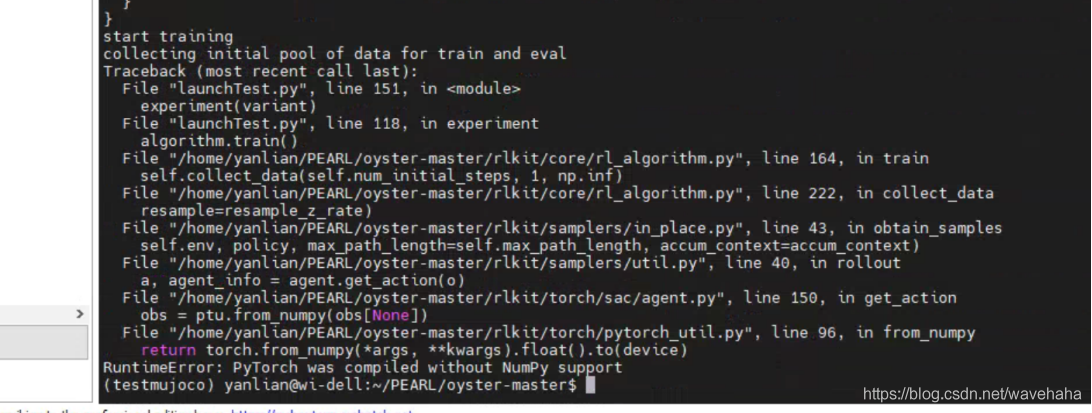
TypeError: Object of type 'bytes' is not JSON serializable
Later, after consulting related materials, I found that many data types of the default encoding function cannot be encoded, so you can write an encoder yourself to inherit jsonencoder, so that it can be encoded.
For example, the above problem is because the json.dumps function found that there are bytes type data in the dictionary, so it cannot be encoded. Just write an encoding class before the encoding function. As long as the data of the bytes type is checked, it will be converted. Into str type.
class MyEncoder(json.JSONEncoder):
def default(self, obj):
if isinstance(obj, bytes):
return str(obj, encoding='utf-8');
return json.JSONEncoder.default(self, obj)
This solved the problem.
Later, similar problems were found during encoding:
TypeError: Object of type 'ndarray' is not JSON serializable
This is the same processing method. When the ndarray data is checked, it is converted into list data:
class MyEncoder(json.JSONEncoder):
def default(self, obj):
if isinstance(obj, np.ndarray):
return obj.tolist()
elif isinstance(obj, bytes):
return str(obj, encoding='utf-8');
return json.JSONEncoder.default(self, obj)
In this way, the data is encoded.
Put the final code for your reference:
import scipy.io as sio
import os
import json
import numpy as np
load_fn = '2%.mat'
load_data = sio.loadmat(load_fn)
print(load_data.keys())
class MyEncoder(json.JSONEncoder):
def default(self, obj):
if isinstance(obj, np.ndarray):
return obj.tolist()
elif isinstance(obj, bytes):
return str(obj, encoding='utf-8');
return json.JSONEncoder.default(self, obj)
save_fn = os.path.splitext(load_fn)[0] + '.json'
file = open(save_fn,'w',encoding='utf-8');
file.write(json.dumps(load_data,cls=MyEncoder,indent=4))
file.close()