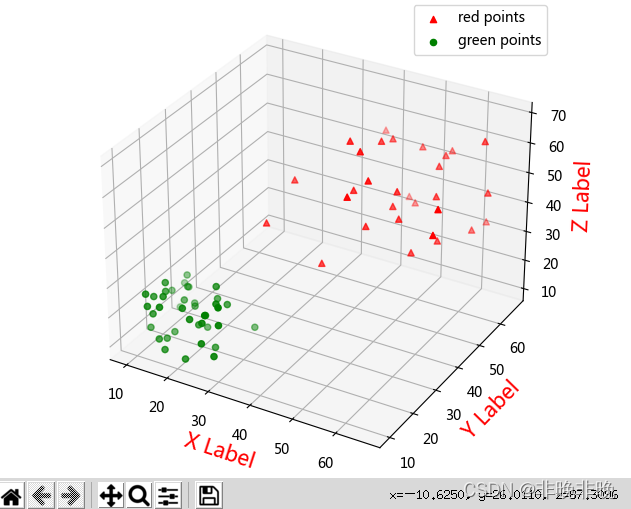
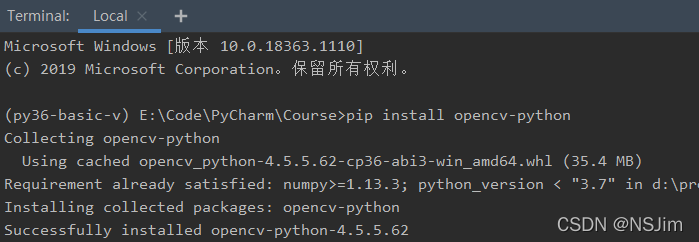
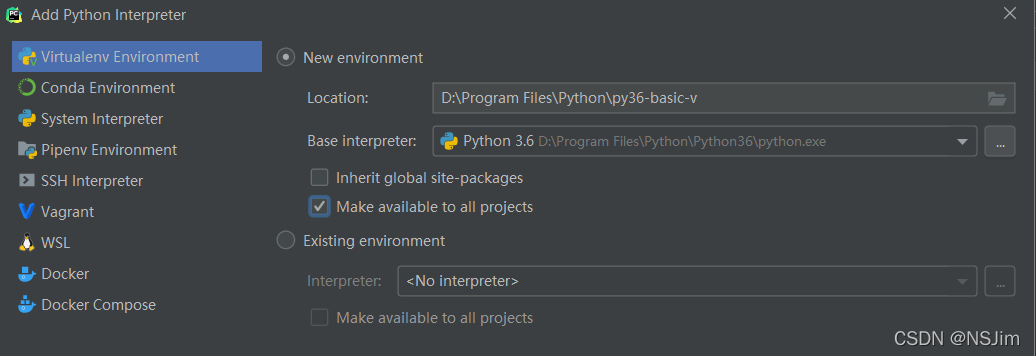
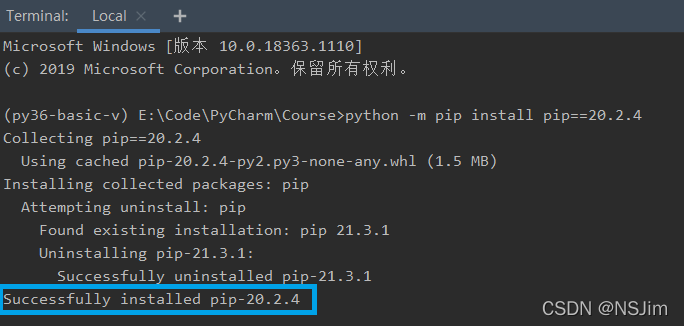
solve the problem
No handles with labels found to put in legend.
Traceback (most recent call last):
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\backends\backend_qt5.py", line 508, in _draw_idle
self.draw()
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\backends\backend_agg.py", line 388, in draw
self.figure.draw(self.renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 38, in draw_wrapper
return draw(artist, renderer, *args, **kwargs)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\figure.py", line 1709, in draw
renderer, self, artists, self.suppressComposite)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\image.py", line 135, in _draw_list_compositing_images
a.draw(renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 38, in draw_wrapper
return draw(artist, renderer, *args, **kwargs)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\axes\_base.py", line 2647, in draw
mimage._draw_list_compositing_images(renderer, self, artists)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\image.py", line 135, in _draw_list_compositing_images
a.draw(renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 38, in draw_wrapper
return draw(artist, renderer, *args, **kwargs)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 670, in draw
bbox, info, descent = textobj._get_layout(renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 276, in _get_layout
key = self.get_prop_tup(renderer=renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 831, in get_prop_tup
x, y = self.get_unitless_position()
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 813, in get_unitless_position
x = float(self.convert_xunits(self._x))
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 180, in convert_xunits
return ax.xaxis.convert_units(x)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\axis.py", line 1553, in convert_units
f'units: {x!r}') from e
matplotlib.units.ConversionError: Failed to convert value(s) to axis units: 'LiR'
Traceback (most recent call last):
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\axis.py", line 1550, in convert_units
ret = self.converter.convert(x, self.units, self)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\category.py", line 52, in convert
'Missing category information for StrCategoryConverter; '
ValueError: Missing category information for StrCategoryConverter; this might be caused by unintendedly mixing categorical and numeric data
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\backends\backend_qt5.py", line 508, in _draw_idle
self.draw()
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\backends\backend_agg.py", line 388, in draw
self.figure.draw(self.renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 38, in draw_wrapper
return draw(artist, renderer, *args, **kwargs)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\figure.py", line 1709, in draw
renderer, self, artists, self.suppressComposite)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\image.py", line 135, in _draw_list_compositing_images
a.draw(renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 38, in draw_wrapper
return draw(artist, renderer, *args, **kwargs)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\axes\_base.py", line 2647, in draw
mimage._draw_list_compositing_images(renderer, self, artists)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\image.py", line 135, in _draw_list_compositing_images
a.draw(renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 38, in draw_wrapper
return draw(artist, renderer, *args, **kwargs)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 670, in draw
bbox, info, descent = textobj._get_layout(renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 276, in _get_layout
key = self.get_prop_tup(renderer=renderer)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 831, in get_prop_tup
x, y = self.get_unitless_position()
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\text.py", line 813, in get_unitless_position
x = float(self.convert_xunits(self._x))
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\artist.py", line 180, in convert_xunits
return ax.xaxis.convert_units(x)
File "D:\ProgramData\Anaconda3\lib\site-packages\matplotlib\axis.py", line 1553, in convert_units
f'units: {x!r}') from e
matplotlib.units.ConversionError: Failed to convert value(s) to axis units: 'LiR'
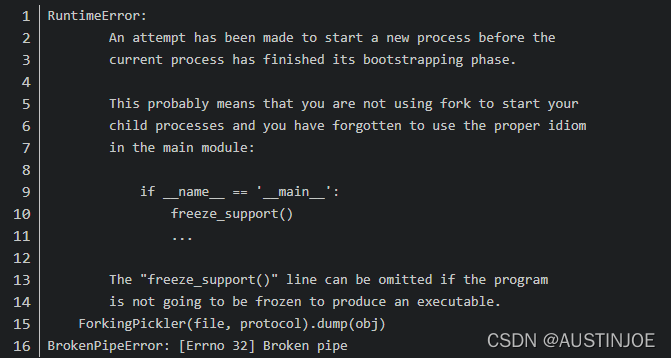
Error:
matplotlib.units.Conversion error: failed to convert value to axis unit: ‘LiR’
Solution:
matplotlib version is low, update the matplotlib library to version 3.3.2 or higher!
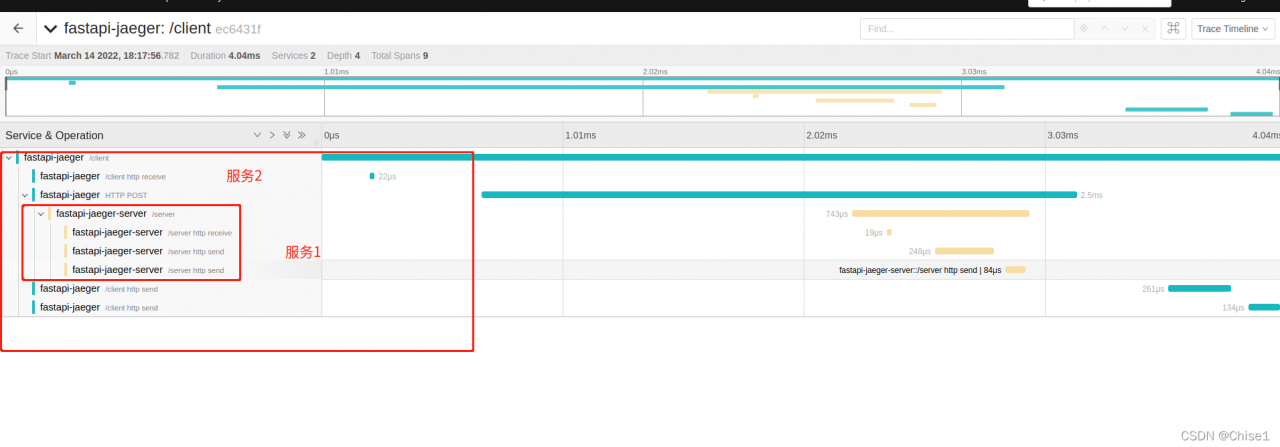 if you have any questions, please correct them.
if you have any questions, please correct them.