When Linux server cross compiles opencv, cmake reports an error
cmake: symbol lookup error: cmake: undefined symbol: _ZN4Json5ValueC1ERKSs
Version information:
opencv is 4.5
cmake uses 3.4
Solution:
update cmake to 3.16.4
When Linux server cross compiles opencv, cmake reports an error
cmake: symbol lookup error: cmake: undefined symbol: _ZN4Json5ValueC1ERKSs
Version information:
opencv is 4.5
cmake uses 3.4
Solution:
update cmake to 3.16.4
1. preface
After using vscode to install the plug-in: glsl lint, there is an error every time you start vscode:
glsl lint: failed to spawn ‘glslangvalidator’ binary Error: spawn glslangValidator. Exe enoent
and vscode will not be automatically colored and highlighted every time the shader program is opened. At this time, I reinstall the glsl lint plug-in. It’s troublesome
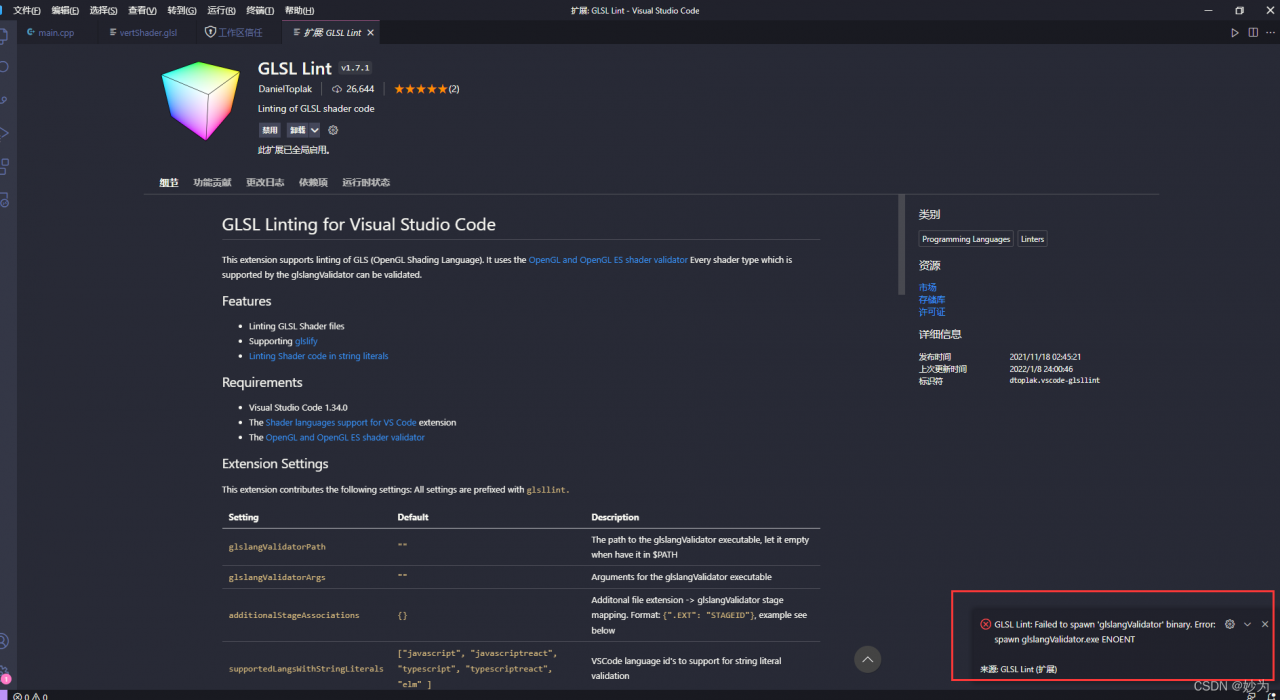
glslangValidator is a custom GLSL reference compiler from the Khronos Group, the official OpenGL standard customizer. The command line compilation mode facilitates users to test glsl syntax directly and bypass the C/C++ related dependency libraries compilation, and there is no need to write a lot of initialization code in the main file. This can save a lot of time in practical testing. In many websites such as shadertoy, the glsl code written by users can be compiled and run directly in the main function, which requires a convenient syntax check, and the shadertoy website also provides a simple syntax check. However, glslang is a standard custom official compiler, so the compiled glsl code can be compiled correctly on all kinds of drivers that support GLSL syntax.
2. Solution steps
1. Open vscode settings
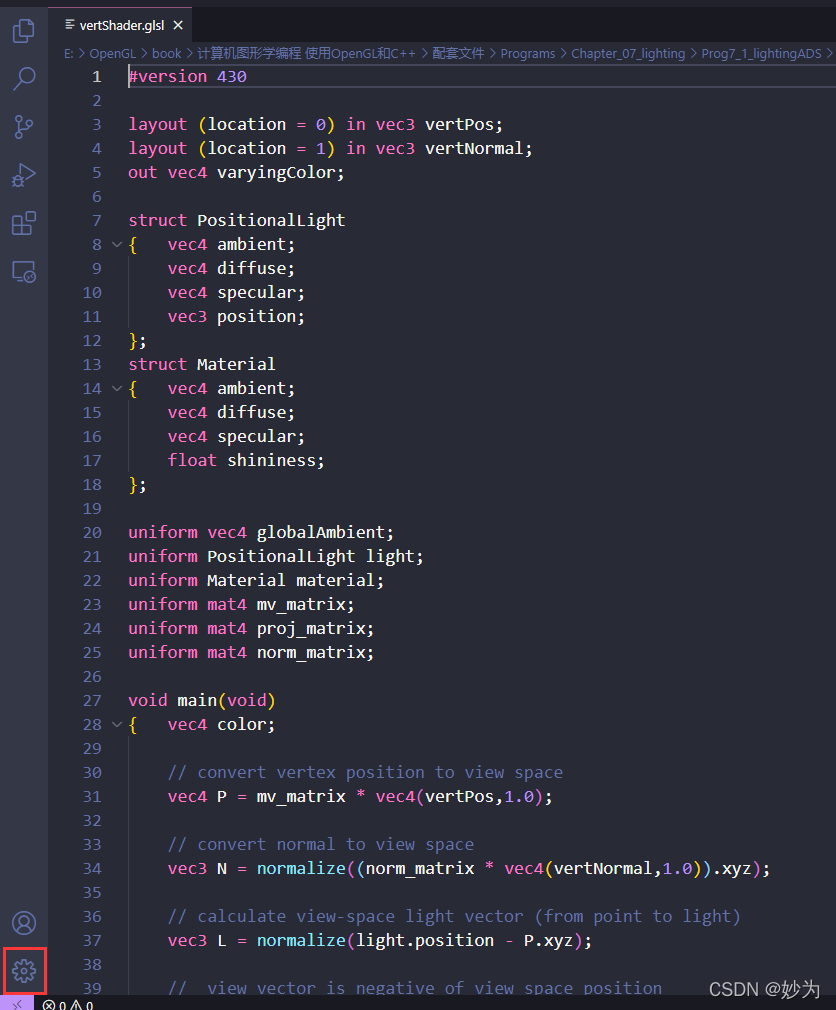
2. Search “security.workspace.trust”
Enter “security.workspace.trust” in “search settings” (just enter “security”)
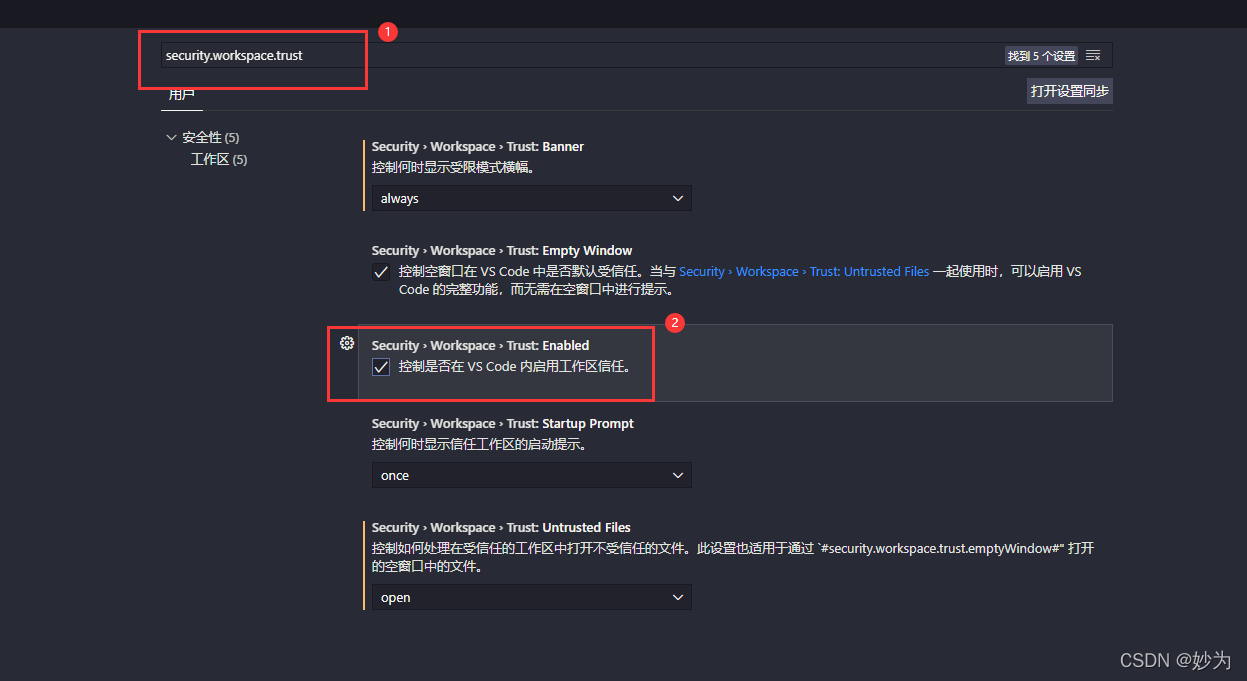
3. Cancel ssecurity.workspace.trust:Enabled
Remove the hook in front of “security.workspace.trust:Enabled”
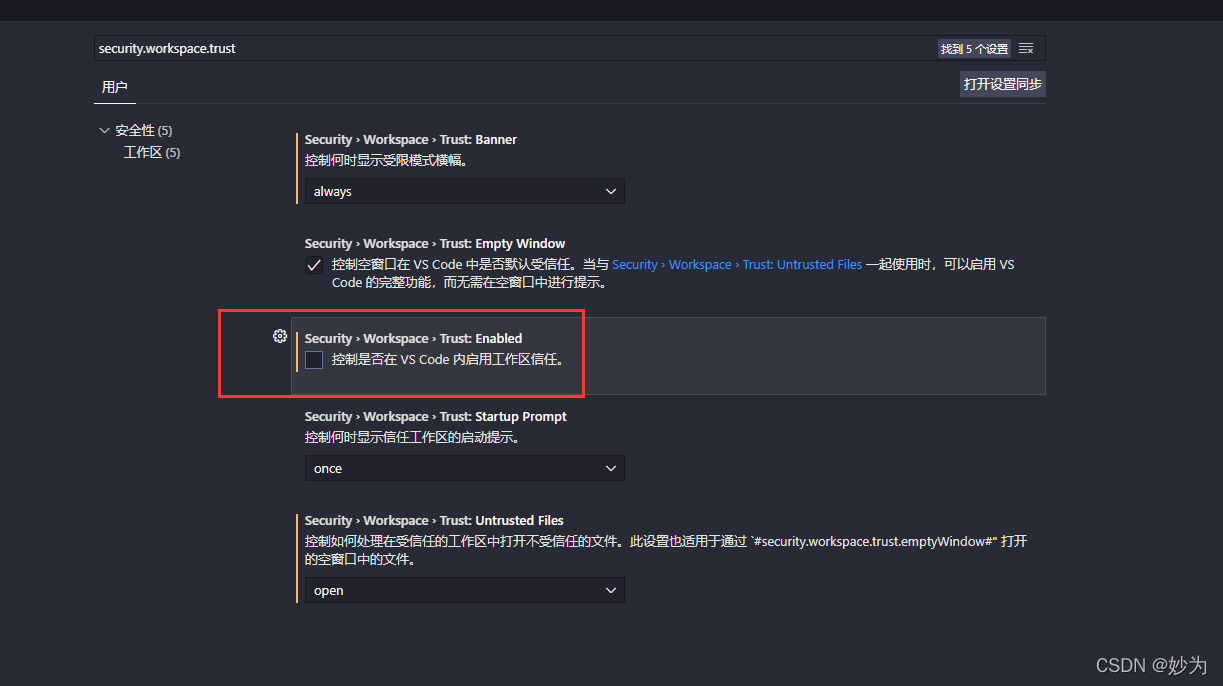
Just restart vscode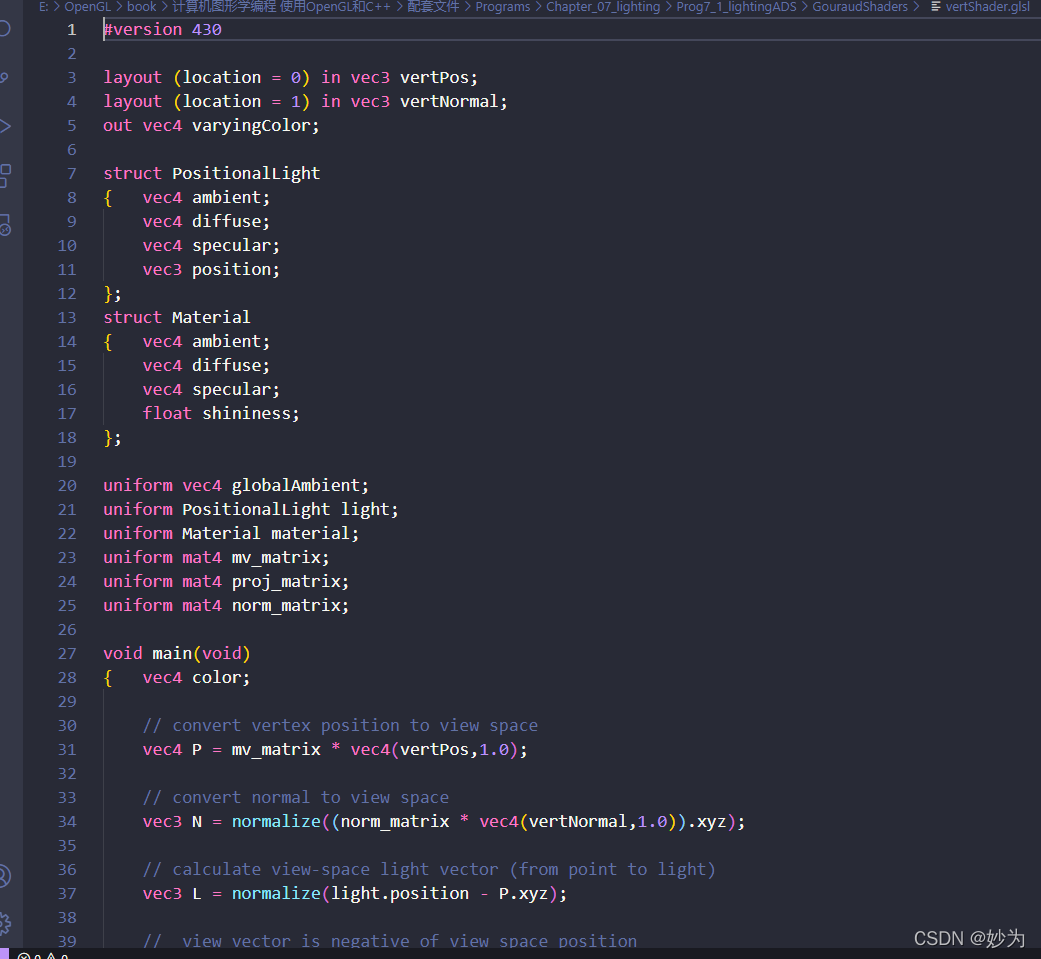
The following problems are encountered when compiling vins-mono with catkin,
In file included from /usr/local/include/ceres/internal/parameter_dims.h:37,
from /usr/local/include/ceres/internal/autodiff.h:151,
from /usr/local/include/ceres/autodiff_cost_function.h:130,
from /usr/local/include/ceres/ceres.h:37,
from /home/matthew/projects/vinsmono/src/VINS-Mono/camera_model/src/calib/CameraCalibration.cc:20:
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:64:21: error: ‘integer_sequence’ is not a member of ‘std’
64 | struct SumImpl<std::integer_sequence<T, N, Ns...>> {
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:64:21: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:64:46: error: wrong number of template arguments (3, should be 1)
64 | struct SumImpl<std::integer_sequence<T, N, Ns...>> {
| ^~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:60:8: note: provided for ‘template<class Seq> struct ceres::internal::SumImpl’
60 | struct SumImpl;
| ^~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:64:49: error: expected unqualified-id before ‘>’ token
64 | struct SumImpl<std::integer_sequence<T, N, Ns...>> {
| ^~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:71:21: error: ‘integer_sequence’ is not a member of ‘std’
71 | struct SumImpl<std::integer_sequence<T, N1, N2, Ns...>> {
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:71:21: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:71:51: error: wrong number of template arguments (4, should be 1)
71 | struct SumImpl<std::integer_sequence<T, N1, N2, Ns...>> {
| ^~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:60:8: note: provided for ‘template<class Seq> struct ceres::internal::SumImpl’
60 | struct SumImpl;
| ^~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:71:54: error: expected unqualified-id before ‘>’ token
71 | struct SumImpl<std::integer_sequence<T, N1, N2, Ns...>> {
| ^~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:78:21: error: ‘integer_sequence’ is not a member of ‘std’
78 | struct SumImpl<std::integer_sequence<T, N1, N2, N3, N4, Ns...>> {
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:78:21: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:78:59: error: wrong number of template arguments (6, should be 1)
78 | struct SumImpl<std::integer_sequence<T, N1, N2, N3, N4, Ns...>> {
| ^~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:60:8: note: provided for ‘template<class Seq> struct ceres::internal::SumImpl’
60 | struct SumImpl;
| ^~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:78:62: error: expected unqualified-id before ‘>’ token
78 | struct SumImpl<std::integer_sequence<T, N1, N2, N3, N4, Ns...>> {
| ^~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:85:21: error: ‘integer_sequence’ is not a member of ‘std’
85 | struct SumImpl<std::integer_sequence<T, N>> {
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:85:21: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:85:41: error: wrong number of template arguments (2, should be 1)
85 | struct SumImpl<std::integer_sequence<T, N>> {
| ^
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:60:8: note: provided for ‘template<class Seq> struct ceres::internal::SumImpl’
60 | struct SumImpl;
| ^~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:85:42: error: expected unqualified-id before ‘>’ token
85 | struct SumImpl<std::integer_sequence<T, N>> {
| ^~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:91:21: error: ‘integer_sequence’ is not a member of ‘std’
91 | struct SumImpl<std::integer_sequence<T>> {
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:91:21: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:91:38: error: template argument 1 is invalid
91 | struct SumImpl<std::integer_sequence<T>> {
| ^
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:91:39: error: expected unqualified-id before ‘>’ token
91 | struct SumImpl<std::integer_sequence<T>> {
| ^~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:135:31: error: ‘integer_sequence’ is not a member of ‘std’
135 | std::integer_sequence<T, N, Ns...>,
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:135:31: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:135:59: error: template argument 3 is invalid
135 | std::integer_sequence<T, N, Ns...>,
| ^
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:135:59: error: type/value mismatch at argument 4 in template parameter list for ‘template<class T, T Sum, class SeqIn, class SeqOut> struct ceres::internal::ExclusiveScanImpl’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:135:59: note: expected a type, got ‘N’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:146:39: error: ‘integer_sequence’ is not a member of ‘std’
146 | struct ExclusiveScanImpl<T, Sum, std::integer_sequence<T>, SeqOut> {
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:146:39: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:146:57: error: wrong number of template arguments (3, should be 4)
146 | struct ExclusiveScanImpl<T, Sum, std::integer_sequence<T>, SeqOut> {
| ^
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:130:8: note: provided for ‘template<class T, T Sum, class SeqIn, class SeqOut> struct ceres::internal::ExclusiveScanImpl’
130 | struct ExclusiveScanImpl;
| ^~~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:160:53: error: ‘integer_sequence’ is not a member of ‘std’
160 | typename ExclusiveScanImpl<T, T(0), Seq, std::integer_sequence<T>>::Type;
| ^~~~~~~~~~~~~~~~
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:160:53: error: ‘integer_sequence’ is not a member of ‘std’
/usr/local/include/ceres/internal/integer_sequence_algorithm.h:160:70: error: template argument 4 is invalid
160 | typename ExclusiveScanImpl<T, T(0), Seq, std::integer_sequence<T>>::Type;
After checking, it is generally caused by the incompatibility between Ceres-solver and eigen3. You can’t use the latest version of Ceres when running vins-mono.
I had no choice but to uninstall the previous version 2.0.0 first.
sudo rm -r /usr/local/lib/cmake/Ceres
sudo rm -rf /usr/local/include/ceres /usr/local/lib/libceres.a
sudo rm -r /usr/local/share/CeresThen download version 1.14.0 here,
http://ceres-solver.org/ceres-solver-1.14.0.tar.gz
wget ceres-solver.org/ceres-solver-1.14.0.tar.gz
https://ceres-solver.googlesource.com/ceres-solver
cd ceres-solver-1.14.0
mkdir build
cd build
cmake ..
make -j4
make test
sudo make install
Compile ceres_curve_fiiting
CD to ceres_curve_fiiting folder
mkdir build
cd build
cmake ..
make
./curve_fitting
Compilation finished!
First, check whether more than half of the servers start zookeeper. If yes, use the JPS command and find that the quorumpeermain main class is not started
[atguigu@Hadoop103 zookeeper-3.5.7]$ jps
14850 JpsThe most likely cause: Zookeeper decompression path in the conf folder zoo.cfg (I am here after changing the name) configuration when adding content after the addition of spaces and the creation of myid up and down there are empty lines or left and right spaces, enter the file deleted, and then check the jps
#######################cluster##########################
server.2=hadoop102:2888:3888
server.3=hadoop103:2888:3888
server.4=hadoop104:2888:3888Problem description
Android studio 4.0.1 in APP/build Gradle declares to use appcompat-v7, and the compilation times the following errors:
Android resource linking failed
Output: D:\code\demo\app\build\intermediates\incremental\mergeDebugResources\merged.dir\values-v28\values-v28.xml:7: error: resource android:attr/dialogCornerRadius not found.
D:\code\demo\app\build\intermediates\incremental\mergeDebugResources\merged.dir\values-v28\values-v28.xml:11: error: resource android:attr/dialogCornerRadius not found.
D:\code\demo\app\build\intermediates\incremental\mergeDebugResources\merged.dir\values\values.xml:1086: error: resource android:attr/fontVariationSettings not found.
D:\code\demo\app\build\intermediates\incremental\mergeDebugResources\merged.dir\values\values.xml:1087: error: resource android:attr/ttcIndex not found.
error: failed linking references.
Command: D:\gradle\caches\transforms-1\files-1.1\aapt2-3.2.1-4818971-windows.jar\67766513f93fbda68bd5705c60a70b26\aapt2-3.2.1-4818971-windows\aapt2.exe link -I\
D:\Android\SDK\platforms\android-27\android.jar\
--manifest\
D:\code\demo\app\build\intermediates\merged_manifests\debug\processDebugManifest\merged\AndroidManifest.xml\
-o\
D:\code\demo\app\build\intermediates\processed_res\debug\processDebugResources\out\resources-debug.ap_\
-R\
@D:\code\demo\app\build\intermediates\incremental\processDebugResources\resources-list-for-resources-debug.ap_.txt\
--auto-add-overlay\
--java\
D:\code\demo\app\build\generated\not_namespaced_r_class_sources\debug\processDebugResources\r\
--custom-package\
com.demo\
-0\
apk\
--output-text-symbols\
D:\code\demo\app\build\intermediates\symbols\debug\R.txt\
--no-version-vectors
Daemon: AAPT2 aapt2-3.2.1-4818971-windows Daemon #0Solution:
In the root directory gradle In the properties file, disable Android X:
# android.useAndroidX=true
# android.enableJetifier=trueIt can be solved by rebuilding.
1. Problem
an error is reported when executing the command NPM run serve:
Error: Cannot find module 'postcss-loader'
2. Solution
The most thorough solution is to uninstall the current nodejs version and install the nodejs version consistent with the module version.
If you don’t want to reinstall nodejs, you can use the following methods:
install module:
npm install postcss-loader
If you continue to report errors:
npm ERR! Could not resolve dependency:
npm ERR! postcss-loader@"*" from the root project
npm ERR!
npm ERR! Conflicting peer dependency: [email protected]
npm ERR! node_modules/webpack
npm ERR! peer webpack@"^5.0.0" from [email protected]
npm ERR! node_modules/postcss-loader
npm ERR! postcss-loader@"*" from the root project
npm ERR!
npm ERR! Fix the upstream dependency conflict, or retry
npm ERR! this command with --force, or --legacy-peer-deps
npm ERR! to accept an incorrect (and potentially broken) dependency resolution.
Take the – force or — legacy-peer-deps parameter after the command:
npm install postcss-loader --legacy-peer-deps
Error Messages:
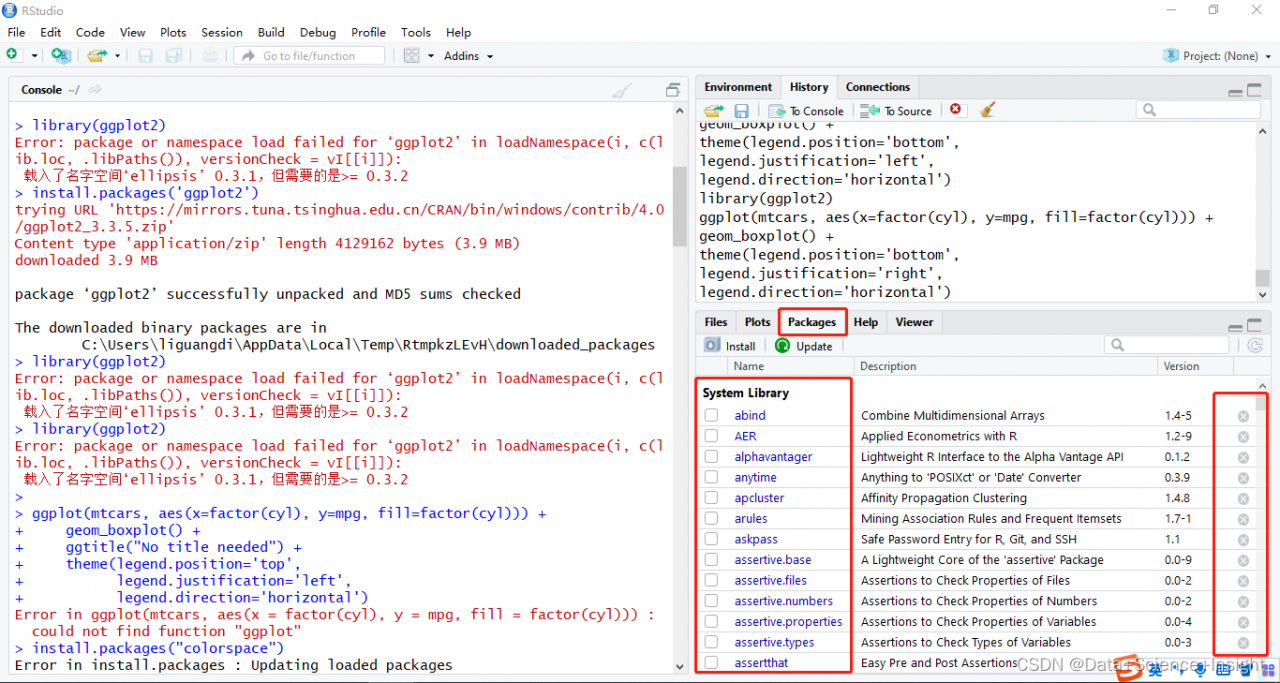
> library(ggplot2)
Error: package or namespace load failed for ‘ggplot2’ in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]):
Loaded namespace ‘ellipsis’ 0.3.1, but what is needed is >= 0.3.2
Solution:
In Rstudio, find the packages module, remove the package that reported the error and then re-install it using install.packages for the corresponding installation.
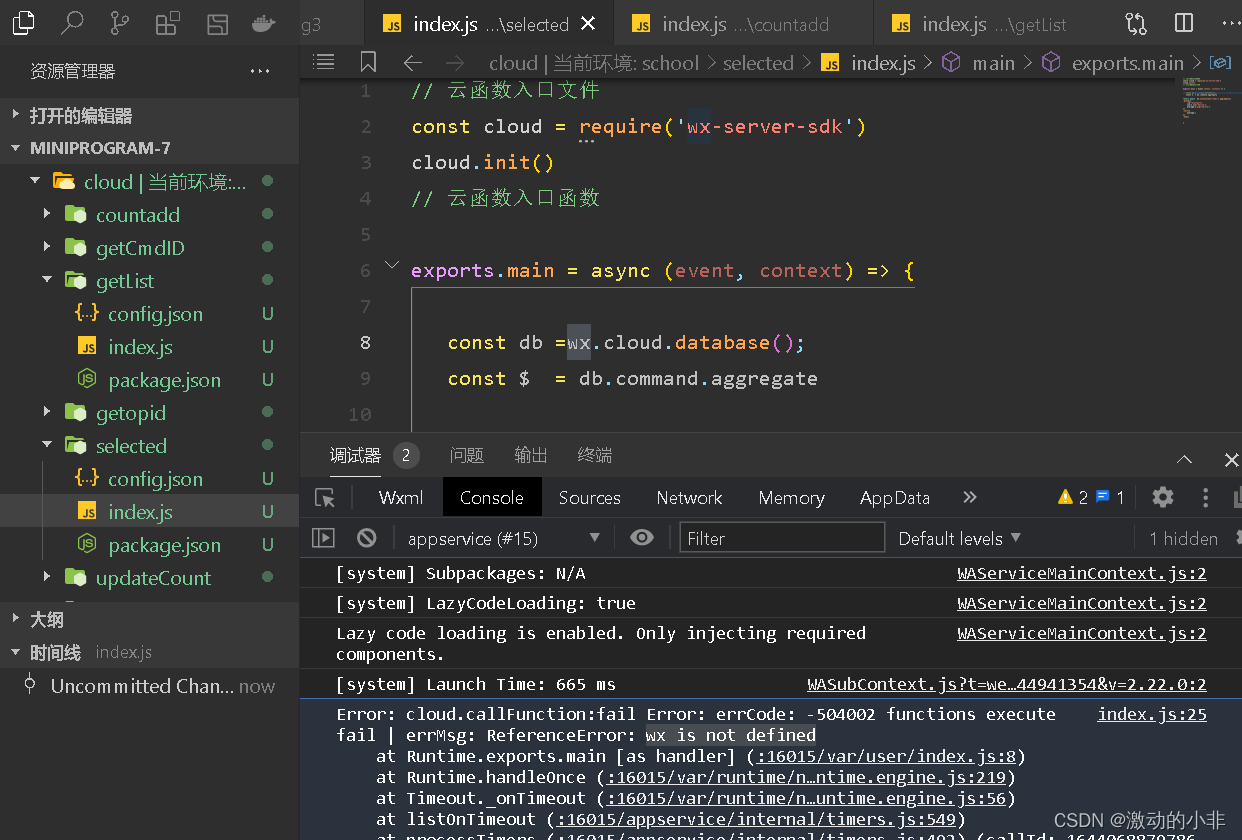
Event: 2022/2/5 21:04
Error reason: in the cloud folder, JS files do not need to use Wx to call functions.
After removing the Wx call, the cloud function is used successfully.
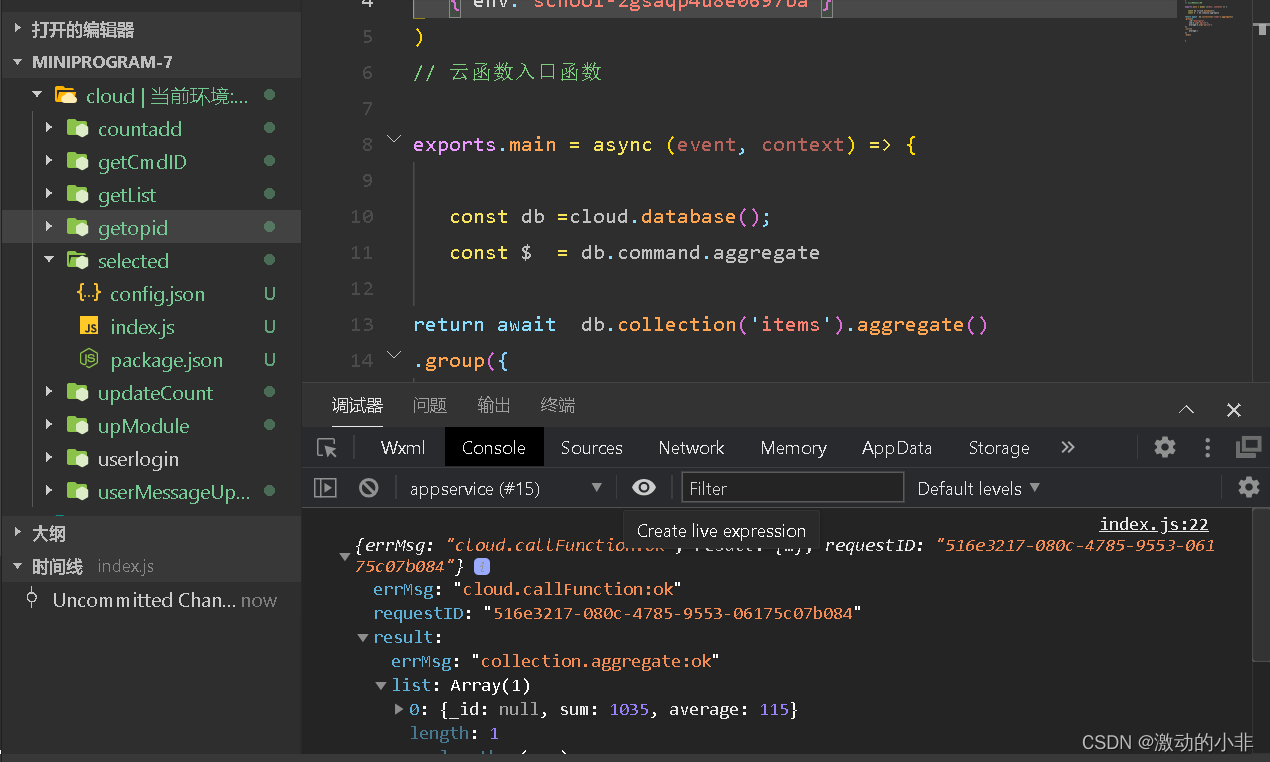
Be careful.
Could not identify launch activity: default activity not found appears in Android studio Error while Launching activity
Check androidmanifest Are the following settings in XML
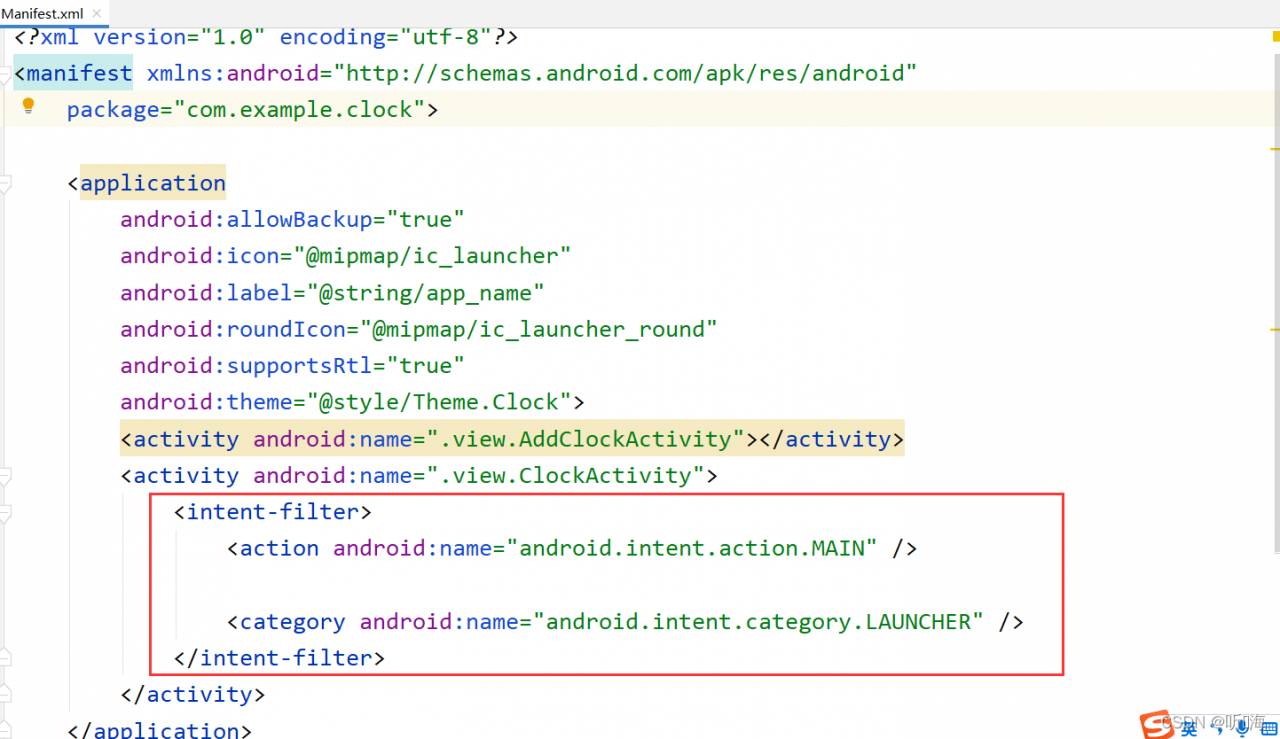
Solution: Configure the following code for the activity to be started
<intent-filter>
<action android:name="android.intent.action.MAIN" />
<category android:name="android.intent.category.LAUNCHER" />
</intent-filter>
process[feature_tracker-2]: started with pid [8459] RLException: Roslaunch got a 'No such file or directory' error while attempting to run: xterm -e gdb --args /home/matthew/projects/vinsmono/devel/lib/vins_estimator/vins_estimator __name:=vins_estimator __log:=/home/matthew/.ros/log/5eeae726-84a9-11ec-ad3c-378f34e64432/vins_estimator-3.log Please make sure that all the executables in this command exist and have executable permission. This is often caused by a bad launch-prefix. The traceback for the exception was written to the log file
Solution:
sudo apt-get install xtermOpencv4 error
This is the source code:
#include <opencv2/opencv.hpp>
#include <iostream>
using namespace cv;
int main(int argc, const char* argv[]) {
Mat src, dst;
src = imread("./test.jpg");
//if (src.empty()) {
if (!src.data){
printf("could not load image...\n");
return -1;
}
//namedWindow("input img");
//imshow("input img", src);
//Three for loops, performing operations g_dstImage(i,j) =a*g_srcImage(i,j) + b
for (int y = 0; y < src.rows; y++)
{
for (int x = 0; x < src.cols; x++)
{
for (int c = 0; c < 3; c++)
{
//g_dstImage.at<Vec3b>(y, x)[c] = saturate_cast<uchar>((g_nContrastValue*0.01)*(g_srcImage.at<Vec3b>(y, x)[c]) + g_nBrightValue);
dst.at<Vec3b>(y, x)[c] = src.at<Vec3b>(y, x)[c];
}
}
}
namedWindow("output img");
imshow("output img", dst);
waitKey(0);
return 0;
}
Vs compile run error:
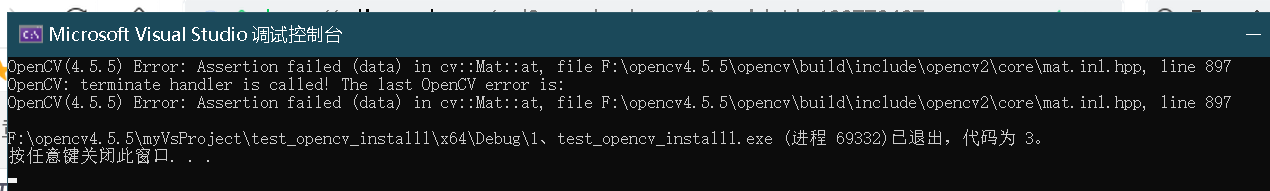
reason
The nonexistent subscript of the access array (because we did not initialize the size and data type of mat array DST in the above code…)
Solution:
Add code: dst = Mat::zeros(src.size(), src.type());
#include <opencv2/opencv.hpp>
#include <iostream>
using namespace cv;
int main(int argc, const char* argv[]) {
Mat src, dst;
src = imread("./test.jpg");
//if (src.empty()) {
if (!src.data){
printf("could not load image...\n");
return -1;
}
namedWindow("input img");
imshow("input img", src);
dst = Mat::zeros(src.size(), src.type());
//Three for loops, performing operations g_dstImage(i,j) =a*g_srcImage(i,j) + b
for (int y = 0; y < src.rows; y++)
{
for (int x = 0; x < src.cols; x++)
{
for (int c = 0; c < 3; c++)
{
//g_dstImage.at<Vec3b>(y, x)[c] = saturate_cast<uchar>((g_nContrastValue*0.01)*(g_srcImage.at<Vec3b>(y, x)[c]) + g_nBrightValue);
dst.at<Vec3b>(y, x)[c] = 255 - src.at<Vec3b>(y, x)[c];
}
}
}
namedWindow("output img");
imshow("output img", dst);
waitKey(0);
return 0;
}
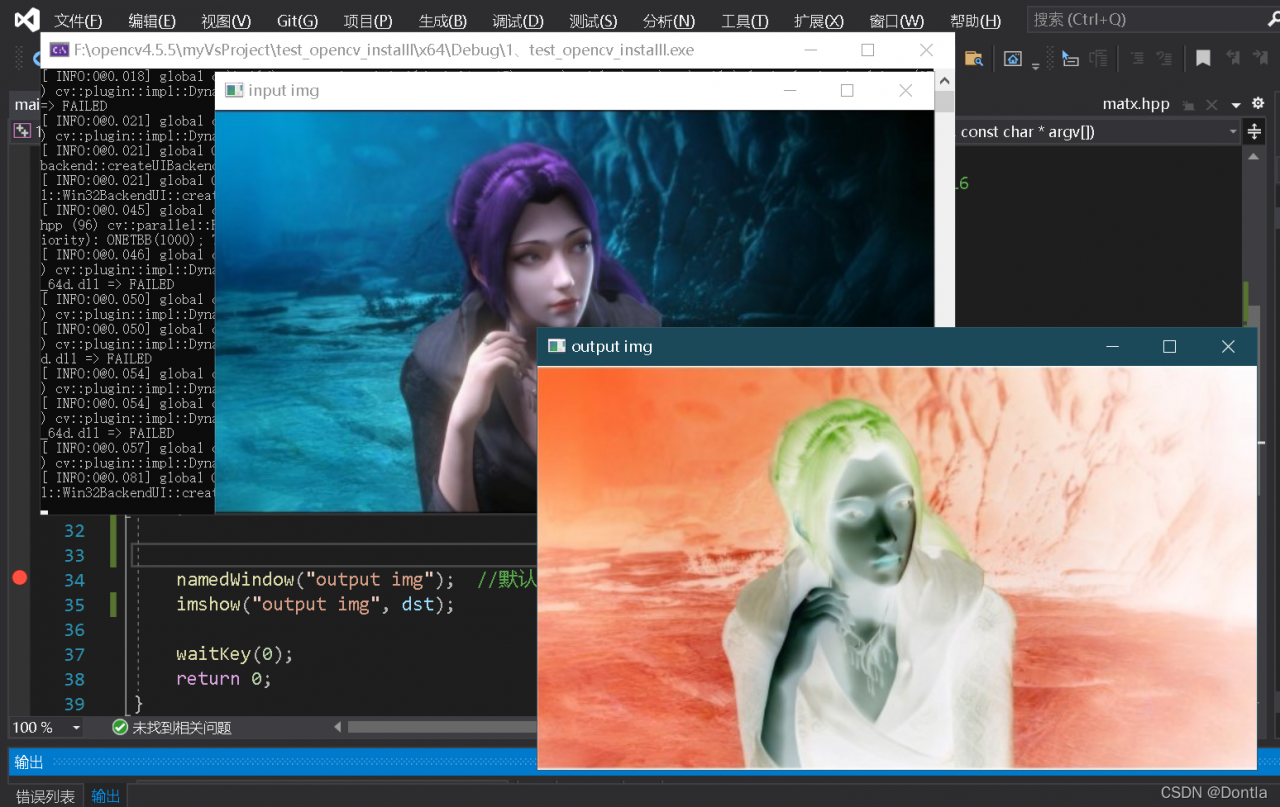
Vs compilation run result:
The v95 version of gbase 8A supports fast replacement of failed nodes through free nodes (omitting the need to find the installation package and check the installation and configuration process). However, free node is only for the scene with VC enabled. For single VC, that is, the mode compatible with V8 version, free node is not supported, so it does not support this node replacement method. The solution is to use a new node.
Error message
version: 9.5.3.22
install prefix: /opt/gbase/10.0.2.103
execute replace node os user: gbase
replaced nodes: ['10.0.2.104']
freenode: ['10.0.2.106']
Error: replace.py(line 1147) -- single vc mode does not support '--freenode'
[gbase@gbase_rh7_003 gcinstall]$
Reason
the cluster is in single VC mode, compatible with V8, and does not support freenode. Look at the bottom of the gcadmin output. There is really no freenode information.
[gbase@gbase_rh7_003 gcinstall]$ gcadmin
CLUSTER STATE: ACTIVE
VIRTUAL CLUSTER MODE: NORMAL
====================================
| GBASE GCWARE CLUSTER INFORMATION |
====================================
| NodeName | IpAddress | gcware |
------------------------------------
| gcware1 | 10.0.2.103 | OPEN |
------------------------------------
====================================================
| GBASE COORDINATOR CLUSTER INFORMATION |
====================================================
| NodeName | IpAddress | gcluster | DataState |
----------------------------------------------------
| coordinator1 | 10.0.2.103 | OPEN | 0 |
----------------------------------------------------
=========================================================================================================
| GBASE DATA CLUSTER INFORMATION |
=========================================================================================================
| NodeName | IpAddress | DistributionId | gnode | syncserver | DataState |
---------------------------------------------------------------------------------------------------------
| node1 | 10.0.2.103 | 1 | OPEN | OPEN | 0 |
---------------------------------------------------------------------------------------------------------
| node2 | 10.0.2.104 | 1 | OPEN | OPEN | 0 |
---------------------------------------------------------------------------------------------------------
| node3 | 10.0.2.105 | 1 | OPEN | OPEN | 0 |
---------------------------------------------------------------------------------------------------------
| node4 | 10.0.2.106 | | OPEN | OPEN | 0 |
---------------------------------------------------------------------------------------------------------
[gbase@gbase_rh7_003 gcinstall]$
Solution
adopt the scheme of new machines and replace nodes on the original IP.