Directory)
Common Errors and their possible causes (self-use) GGplot class data manipulation and function class
Update on 2019.05.11
Common Errors and their possible causes (for personal use)
The following are all their actual process met, now collected, easy to learn to use, you have encountered other, I can also collect and update into here ~
Ps: Even if you understand the error message, you may not be able to find out which step is wrong, so don’t spray “the error message has explained blabla or other words”.
Ggplot class
- Q: ggplot2_Error: geom_point requires the following missing desire: y
A: stat = “indentity” or “density”, Q: Error: Discrete value to continuous scale
A: The X or Y axis may be set to discrete, while data is continuous, and vice versa. For example, scale_x_Continuous (blablabla) is set, but the X-axis data is C (” val1 “, “val2”,blabal2), which creates a conflict, so errorxlim Ylim and Coord_cartesian might have the same effect with graphs, but not the same. Coord_cartesian is a local zoom that does not change the dataset, while Xlim Ylim is a filter dataset that does change the dataset.
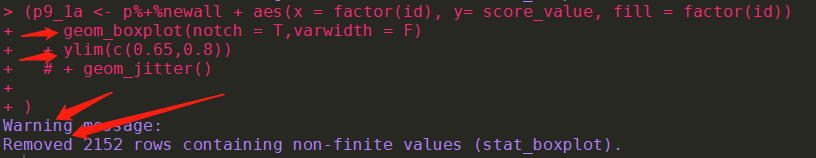
Data manipulation and function classes
- dat[datx > 5, c(1,9)] # only select column 1 and 9
dat[datx > 5, 1:9] # select column 1 to 9. Although these two are different, the tibble of tidyverse bag has the property of data.frame, but it is not completely equal to data.frame:
error:
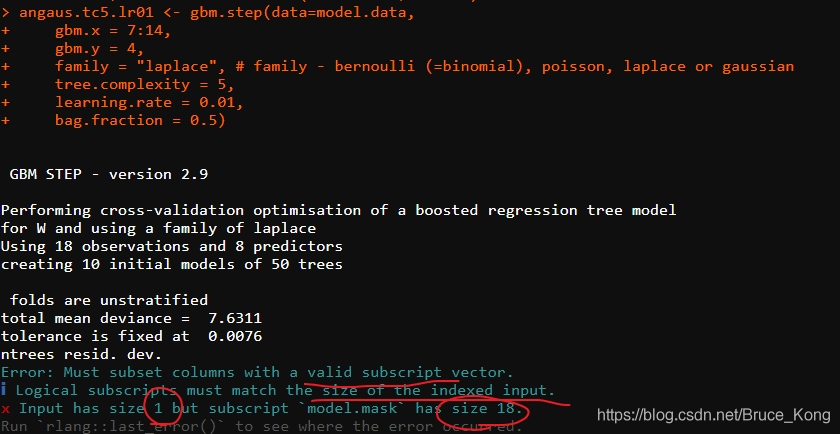
angaus.tc5.lr01 < – GBm. step(Data =model.data, GBM. x = 3:14, GBM. y = 2, family = “Bernoulli”, Tree.com Plexity = 5, learning.rate = 0.01, Bag.Fraction = 0.5)
Error: Must subset columns with a valid subscript vector.
ℹ Logical subscripts Must match the size of the indexed input.
x input from size 1 but subscriptmodel. The mask has size 18.
Runrlang::last_error()to see where the error occurred.
This means that the columns of model.data cannot be recognized. Since model.data is the tidyverse package read_delim, try it and add as.data.frame().
Read More:
- Java common errors < identifier > expected
- Common errors and solutions in MapReduce stage
- SQL Union, union all usage and common errors and Solutions
- Analysis and solution of common errors in ArcEngine development
- Summary of common errors in angularjs
- Common errors and modification methods of findbug
- Common errors in k8s initialization_ [WARNING NumCPU]_ [WARNING SystemVerification]_ WARNING Hostname
- How to Solve Yolov5 Common Error (Three Errors)
- Common errors reported by Eureka in spring cloud
- Common errors and solutions of Qt development application under Ubuntu
- Common solutions to errors in checkout or commit of SVN
- C + + common errors: “error: XXX in namespace ‘STD’ does not name a template type”
- Kafka connection abnormal org.apache.kafka . common.errors.TimeoutException : Failed to update metadata after 60000 ms.
- Error: php56w-common conflicts with php-common-5.3.3-48.el6_ 8.x86_ 64
- Python windows error: [error 3] and common windows error errors
- VTK learning notes: visual model
- Notes on flex RSL application
- ISLR reading notes (3) classification
- R learning notes (1) — ARIMA model
- Learning notes of OpenGL — blending