Error: ‘false’ undeclared (first use in this function)
When you knock the code with DEVC + +, you will report an error to the following program
bool ok(int t){
//Determine whether the tth person's job is assigned or not; if it is not assigned then a[j] = 0, otherwise a[j] = 1.
int i;
for(i=0;i<t;i++)
if(a[i]==a[t])
return false;
return true;
}
analysis:
There are no these keywords in real C. There is no keyword bool in C and early C + +. Bool can be used, but bool is not a built-in type. It is defined through typedef or macros. It is usually defined as int type. Later, the built-in type bool appeared in C + +, and the values can only be true (1) and false (0).
Solution 1:
Change the file type to CPP
Solution 2:
Macro definition of bool:
typedef enum __bool { false = 0, true = 1, } bool;
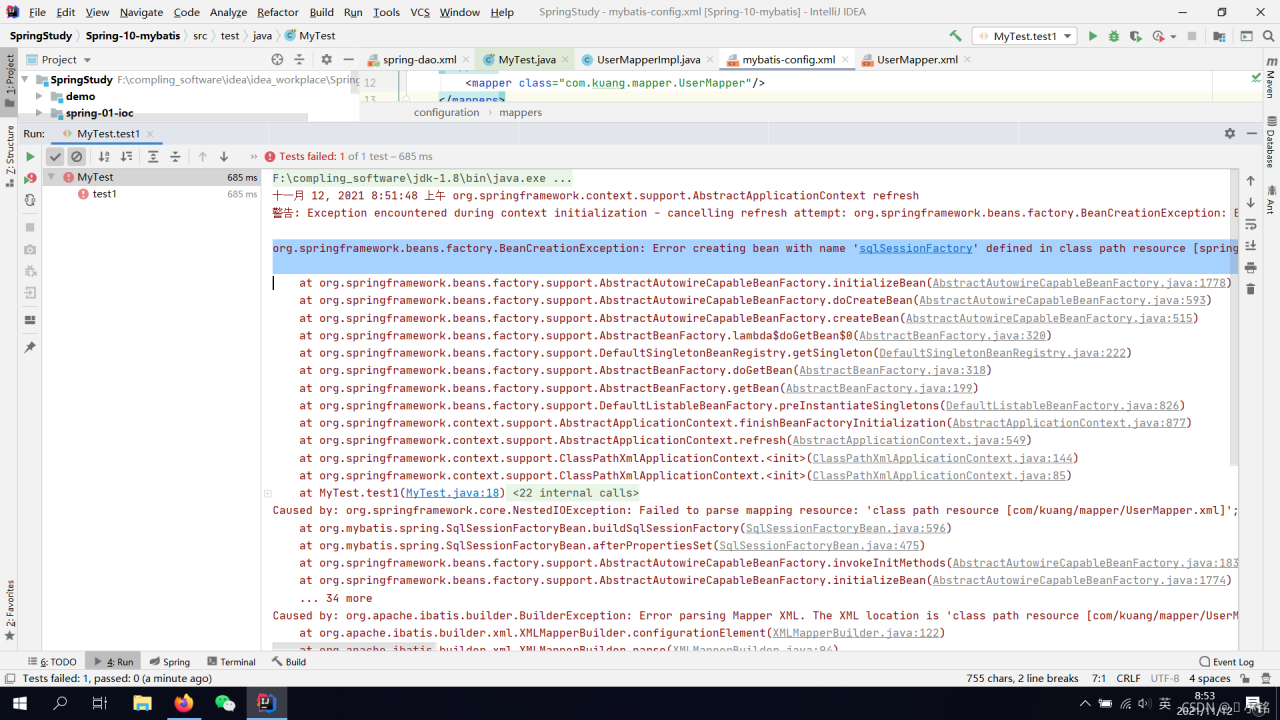 Reason:
Reason: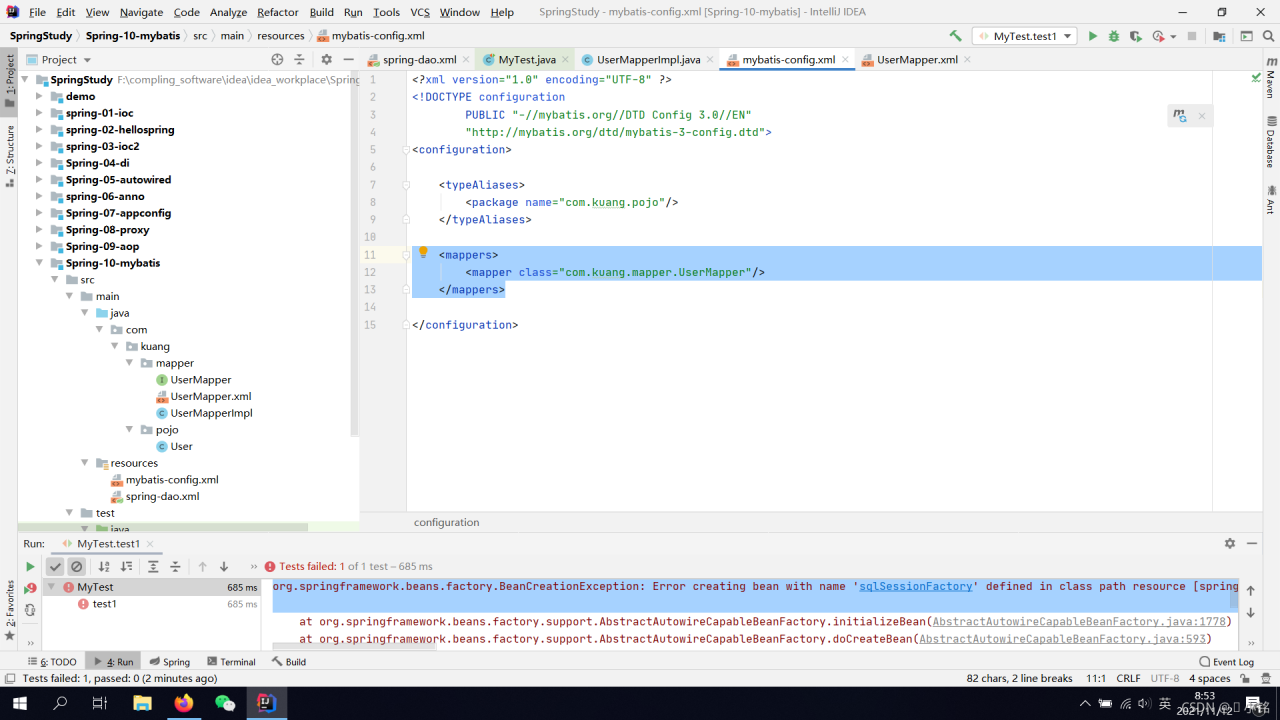
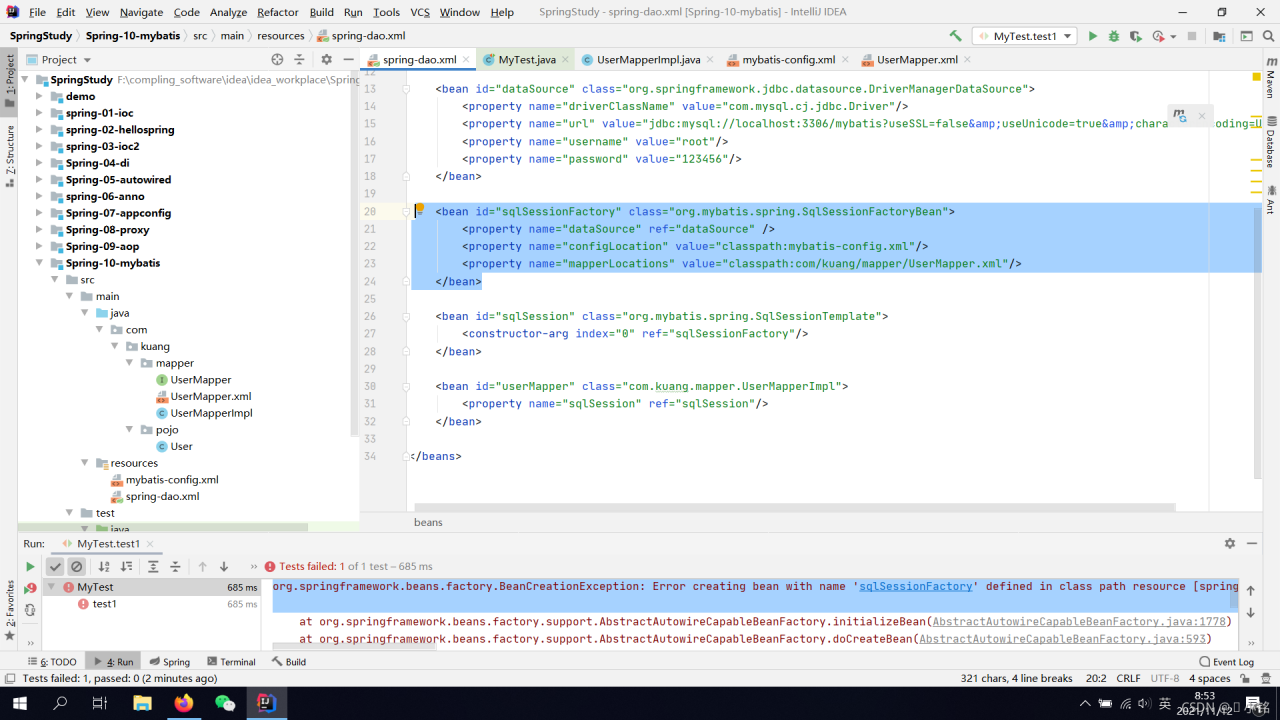 Conflicts with Spring inside
Conflicts with Spring inside