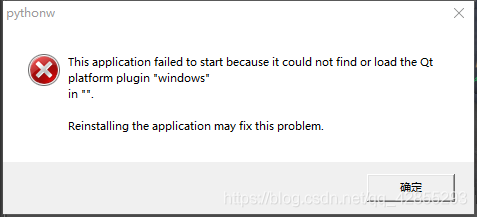
The above is the error message
The solution is
Before instantiating with QApplication() add
QApplication.addLibraryPath(“PySide2 file location” + ‘/PySide2/Plugins/’)
Tag Archives: python
Panda error in modifying line name index does not support mutable operations
During the panda data operation, two tricks appear during expansion:
Using data_PD. Append(), when expanding data in rows, the row names need to be the same in order to realize automatic expansion and use data_ PD. Columns = [], when modifying the row name, slicing is not allowed, and only a list can be created according to the original data length for assignment modification
# Iterate through different individual data and perform data stitching
response_pd = pd.DataFrame() # Create an empty panda
for ii in range(100, 106):
sub = 'Sub%d'%ii
index = np.where((np.array(filter_pd[sub+'_fg'])!=-100)&
(np.array(filter_pd[sub+'_frt'])!=-100))[0]
ii=ii-100
col_index =list(range(2, 7))+list(range(8, 20))+[23+ii*4,21+ii*4,22+ii*4]
# Filter the data in the specified row
temp_pd=filter_pd.iloc[index].iloc[:, col_index]
## Modify the row names, note that the slicing operation is not allowed here, only the original data length, create a list to assign values
temp_pd.columns = list(temp_pd.columns)[:-3]+['first glance', 'first RT', 'last RT']
# Expand the rows, note that you need the same row name to expand the data for the corresponding row
response_pd = response_pd.append(temp_pd,
ignore_index=True)
print('table shape',response_pd.shape)
Python asynchronous co process crawler error: [aiohttp. Client]_ Exceptions: serverdisconnected error: Server Disconnected]
Background description:
I just started to contact crawlers, read online tutorials and began to learn a little bit. All the knowledge points are relatively shallow. If there are better methods, please comment and share.
The initial crawler is very simple: crawl the data list in a web page, and the format returned by the web page is also very simple. It is in the form of a dictionary, which can be directly accessed by saving it as a dictionary with . Json() .
At the beginning of contact with asynchronous collaborative process, after completing the exercise, try to transform the original crawler, resulting in an error.
Initial code:
async def download_page(url):
async with aiohttp.ClientSession() as session:
async with session.get(url) as resp:
await result = resp.text()
async def main(urls):
tasks = []
for url in urls:
tasks.append(asyncio.create_task(download_page(url))) # 我的python版本为3.9.6
await asyncio.await(tasks)
if __name__ == '__main__':
urls = [ url1, url2, …… ]
asyncio.run(main(urls))
This is the most basic asynchronous collaborative process framework. When the amount of data is small, it can basically meet the requirements. However, if the amount of data is slightly large, it will report errors. The error information I collected is as follows:
aiohttp.client_exceptions.ClientOSError: [WinError 64] The specified network name is no longer available.
Task exception was never retrieved
aiohttp.client_exceptions.ClientOSError: [WinError 121] The signal timeout has expired
Task exception was never retrieved
aiohttp.client_exceptions.ServerDisconnectedError: Server disconnected
Task exception was never retrieved
The general error message is that there is a problem with the network request and connection
as a beginner, I didn’t have more in-depth understanding of network connection, nor did I learn more in-depth asynchronous co process operation. Therefore, it is difficult to solve the problem of error reporting.
Solution:
The big problem with the above error reports is that each task creates a session. When too many sessions are created, an error will be reported.
Solution:
Try to create only one session
async def download_page(url,session):
async with session.get(url) as resp:
await result = resp.text()
async def main(urls):
tasks = []
async with aiohttp.ClientSession() as session: # The session will be created in the main function, and the session will be passed to the download_page function as a variable.
for url in urls:
tasks.append(asyncio.create_task(download_page(url,session)))
#My python version is 3.9.6, python version 3.8 and above, if you need to create asynchronous tasks, you need to create them via asyncio.create_task(), otherwise it will run normally but will give a warning message await asyncio.await(tasks)
if __name__ == '__main__':
urls = [ url1, url2, …… ]
asyncio.run(main(urls))
In this way, the problem of connection error can be solved to a certain extent when crawling a large amount of data.
Sentiment: in the process of programming, thinking should be more flexible. A little change may improve the efficiency a lot.
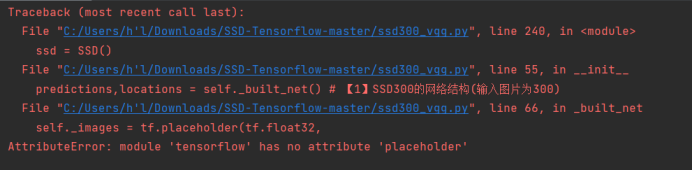
[Solved] Error: AttributeError: module ‘tensorflow‘ has no attribute ‘placeholder‘
Errors are reported as follows:
Solution:
Reason: use tensorflow1 instead of tensorflow2.
Check the tensorflow version information first:
pip list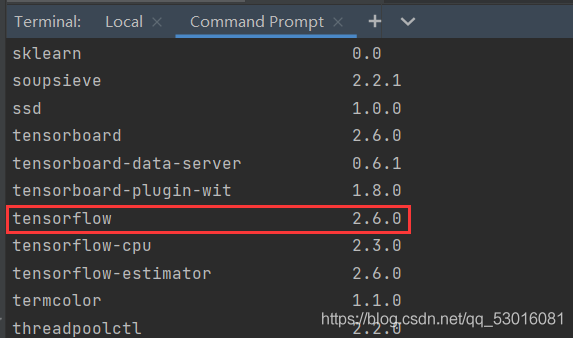
Then add the following code at the beginning of the code:
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()Python read / write file error valueerror: I/O operation on closed file
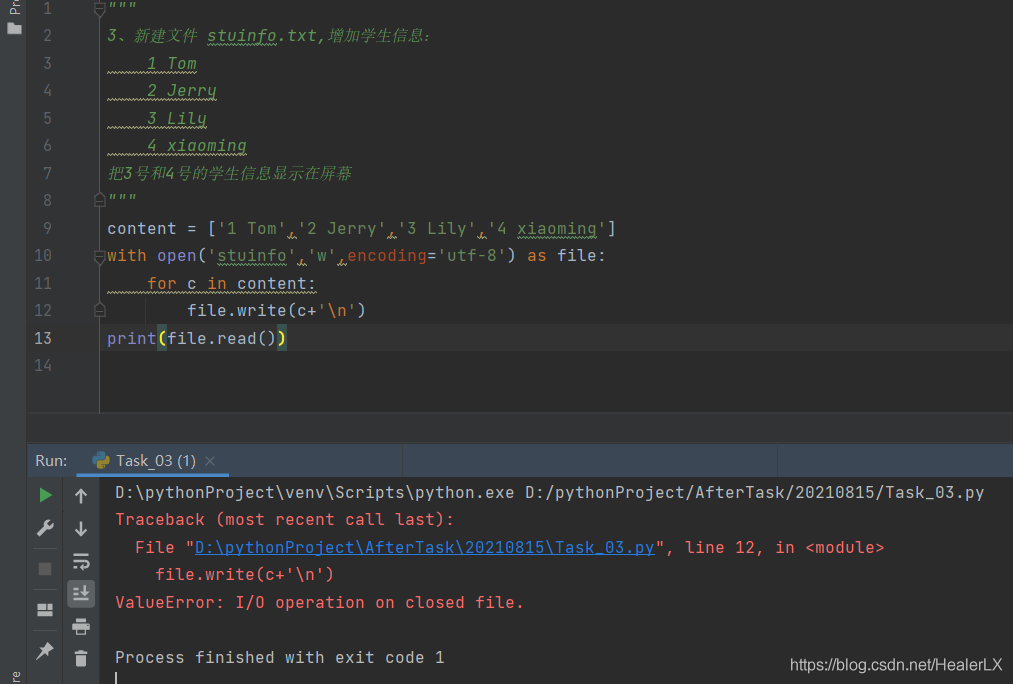
Because with open will turn off file reading and writing by default, you need to open the file content in a read mode before file. Read()
No matching distribution found for exceptions when Python installs docx package
Python installation docx package runs with errors
ERROR: No matching distribution found for exceptions
reason
Docx package is a third-party library developed with Python 2 for Word 2007. With the upgrading of word, especially the upgrading of Python version, it is no longer applicable. You can install a new word processing package to solve the problem.
pip install python-docx
Dm7 Dameng database dmrman reports an error OS_ pipe2_ conn_ Server open failed solution
An error is reported when executing a command in dmrman (DMAP service background starts normally):
RMAN> BACKUP DATABASE '/dm/dmdbms/data/DAMENG/dm.ini';
BACKUP DATABASE '/dm/dmdbms/data/DAMENG/dm.ini';
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[4].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[3].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[2].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[1].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[0].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running, write dmrman info.
EP[0] max_lsn: 155855
BACKUP DATABASE [DAMENG], execute......
os_pipe2_conn_server open failed, name:[/dm/dm_bak/DM_PIPE_DMAP_LSNR_WR], errno:2
CMD END.CODE:[-7109],DESC:[Pipe connect failure]
[-7109]:Pipe connect failure
RMAN>resolvent:
Method (1)
Must go to DM_ Execute dmrman in the home/bin directory. Although the environment variable is configured to recognize the dmrman command, an error will be reported during execution. It is speculated that the files relied on during program execution cannot be recognized correctly. Switch to $DM_ Home/bin solves this problem.
[[email protected] bin]$ pwd
/dm/dmdbms/bin
[[email protected] bin]$ ./dmrman
dmrman V7.6.0.95-Build(2018.09.13-97108)ENT
RMAN> BACKUP DATABASE '/dm/dmdbms/data/DAMENG/dm.ini' FULL BACKUPSET '/dm/dm_bak/db_full_bak_01';
BACKUP DATABASE '/dm/dmdbms/data/DAMENG/dm.ini' FULL BACKUPSET '/dm/dm_bak/db_full_bak_01';
file dm.key not found, use default license!
Global parameter value of RT_HEAP_TARGET is illegal, use min value!
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[4].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[3].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[2].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[1].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running...[0].
checking if the database under system path [/dm/dmdbms/data/DAMENG] is running, write dmrman info.
EP[0] max_lsn: 116978
BACKUP DATABASE [DAMENG], execute......
CMD CHECK LSN......
BACKUP DATABASE [DAMENG], collect dbf......
CMD CHECK ......
DBF BACKUP SUBS......
total 1 packages processed...
total 3 packages processed...
total 4 packages processed...
DBF BACKUP MAIN......
BACKUPSET [/dm/dm_bak/db_full_bak_01] END, CODE [0]......
META GENERATING......
total 5 packages processed...
total 5 packages processed!
CMD END.CODE:[0]
backup successfully!
time used: 7081.941(ms)
RMAN>Method (2)
An error is reported in the log. The pipeline file already exists. An error is reported after deleting the pipeline file OS_ pipe2_ Server open failed, check whether the data is written without permission, modify the permission, and then report an error. The pipeline connection timed out
use./dmrman use_ AP = 2 restore without pipeline DMAP succeeded.
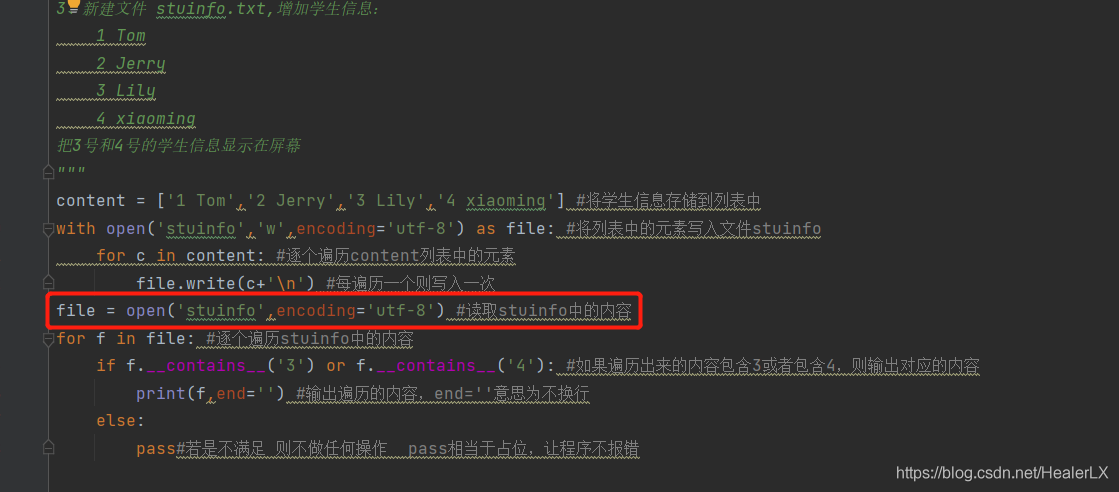
Jenkins reported an error modulenotfounderror: no module named filename solution
1、 If the name of the module is the package name created by yourself, as shown in the picture:
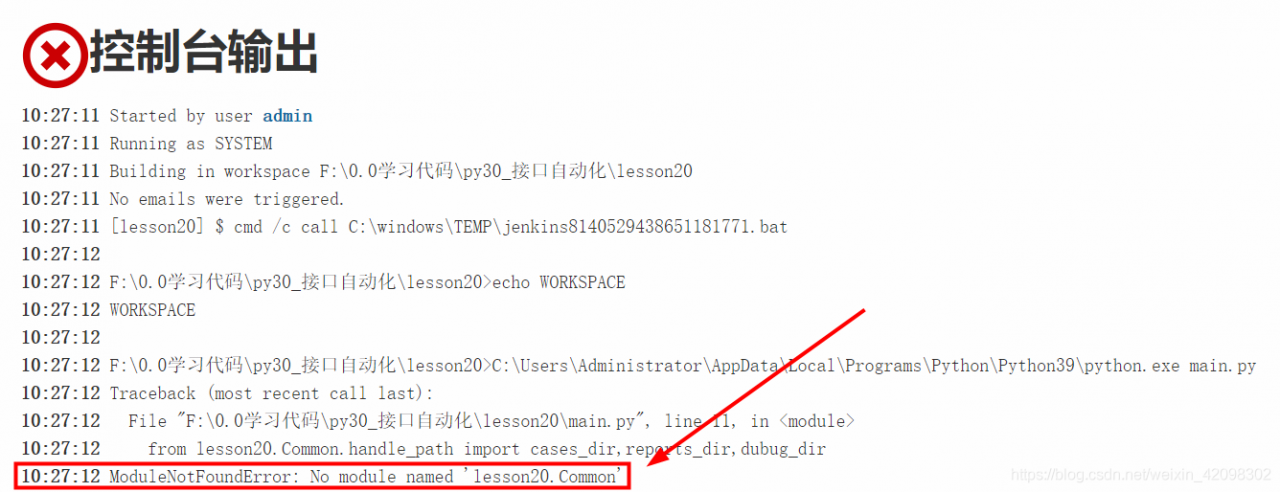
1. Add the following code at the top of the file to be run, and the content in quotation marks is the project path:
import sys
sys.path.append(r"C:\Users\Administrator\XXXProjects\XXX")Note: be sure to write it before all the codes introduced into the module, for example:
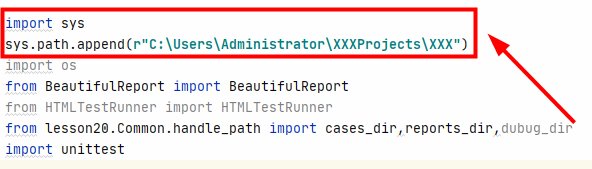
2. Add two lines of code in 1 to all imported files, such as runmethod and get in the above figure_ Data and other files need to be added
3. Another method: create a new file base.py, add the following code, and import it before the codes of all imported modules:
import sys
import os
curPath = os.path.abspath(os.path.dirname(__file__))
rootPath = os.path.abspath(os.path.dirname(curPath) + os.path.sep + ".")
sys.path.append(rootPath)
2、 If the module name is a python module, you need to reinstall it, that is, enter the command PIP install XXX in CMD
Solve the problem of error reporting from scipy.misc import imread & imresize in Python
from scipy.misc import imread& imresize
Problem description solution 1 solution 2
Problem description
from scipy.misc import imread # error
After query, the reason is that the from scipy.misc import imread, imwrite and other methods have been abandoned, and python has encapsulated the imread method in the imageio module
Solution 1
To install the imageio Library:
pip install imageio
Normal use!
import imageio
imageio.imread("xxxx.png")
Solution 2
If the higher version is discarded, we can reduce the SciPy version!
Reduce the SciPy version to 1.2.1
pip install scipy==1.2.1[Solved] Error “incorrect padding” in decoding of Base64 module in Python
Problem description
decodes the field information encoded by Base64, which is normally decoded in the base64 encoding and decoding tool, but b64decode and A2B in the modules Base64 and binascii under python_ Decoding error in Base64 and other methods
the error information is as follows
---------------------------------------------------------------------------
Error Traceback (most recent call last)
<ipython-input-11-787bc11958b4> in get_proxies(urls)
14 try:
---> 15 raw = base64.b64decode(response)
16 except Exception as r:
c:\program files\python3\lib\base64.py in b64decode(s, altchars, validate)
86 raise binascii.Error('Non-base64 digit found')
---> 87 return binascii.a2b_base64(s)
88
Error: Incorrect padding
Solution
Base64 in Python is read in 4, so the field to be decoded should be a multiple of 4, not enough to add ‘=’
# The field a to be decoded is judged, and if it is a multiple of 4, it is unchanged, and if not, how much is missing, how much is made up
a = a + '=' * (4 - len(a) % 4) if len(a) % 4 != 0 else a
Python fbprophet predicts that the code runs with an error
Prophet was successfully installed before, but the following error occurred when the computer was changed. Record it
if Microsoft Visual C + + 14.0 is missing, download a visual studio installer, https://docs.microsoft.com/en-us/visualstudio/install/install-visual-studio?view=vs -In 2019, you can install visual studio 2017
if the error is plot, install plot
If an error is reported, exception ignored in: ‘stanfit4anon_ Model, Download pystan, delete the pystan package in the site packages under CONDA, delete the cache of pystan in appdata \ local \ temp under the user, and then reinstall it.
[Solved] Mindspot error: Error: runtimeerror:_kernel.cc:88 CheckParam] AddN output shape must be equal to input…
Mindspot rewrites withlosscell, traionestepcell interface
Error: runtimeerror:_kernel.cc:88 CheckParam] AddN output shape must be equal to input shape.Trace: In file add_impl.py(272)/ return F.addn((x, y))/
Solution: do not return multiple parameters in the construct method of withlosscell, otherwise an error will be reported in the gradoperation in the construct of traionestepcell.