import statsmodels. API as SM reports the following errors:
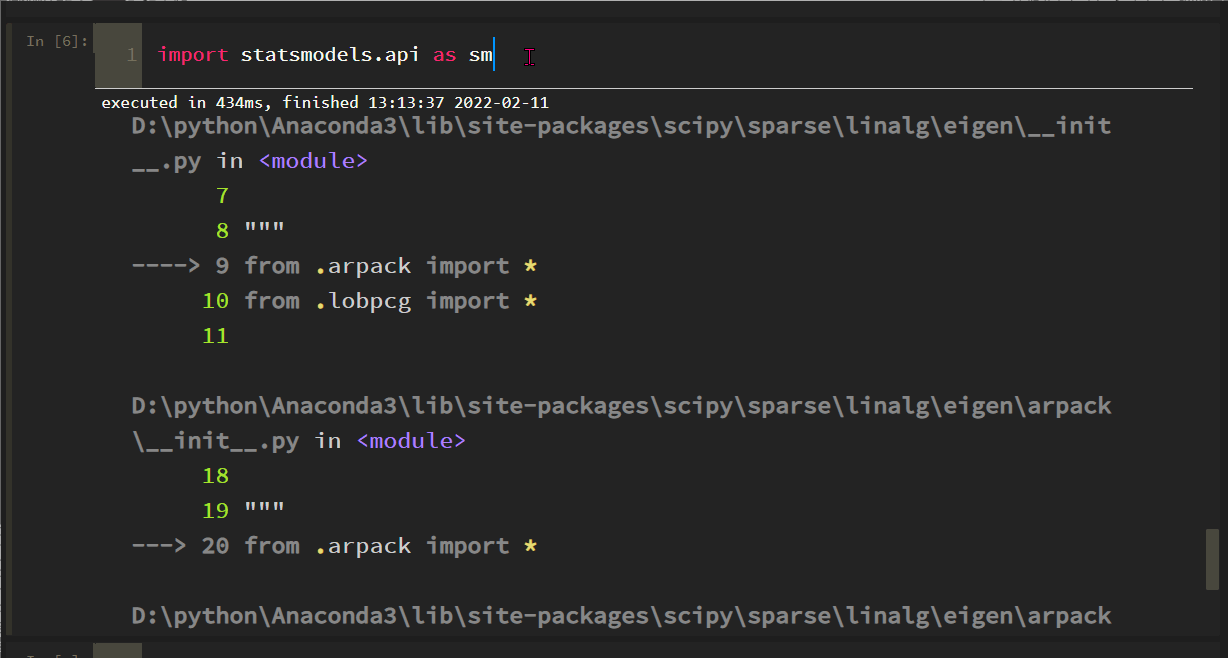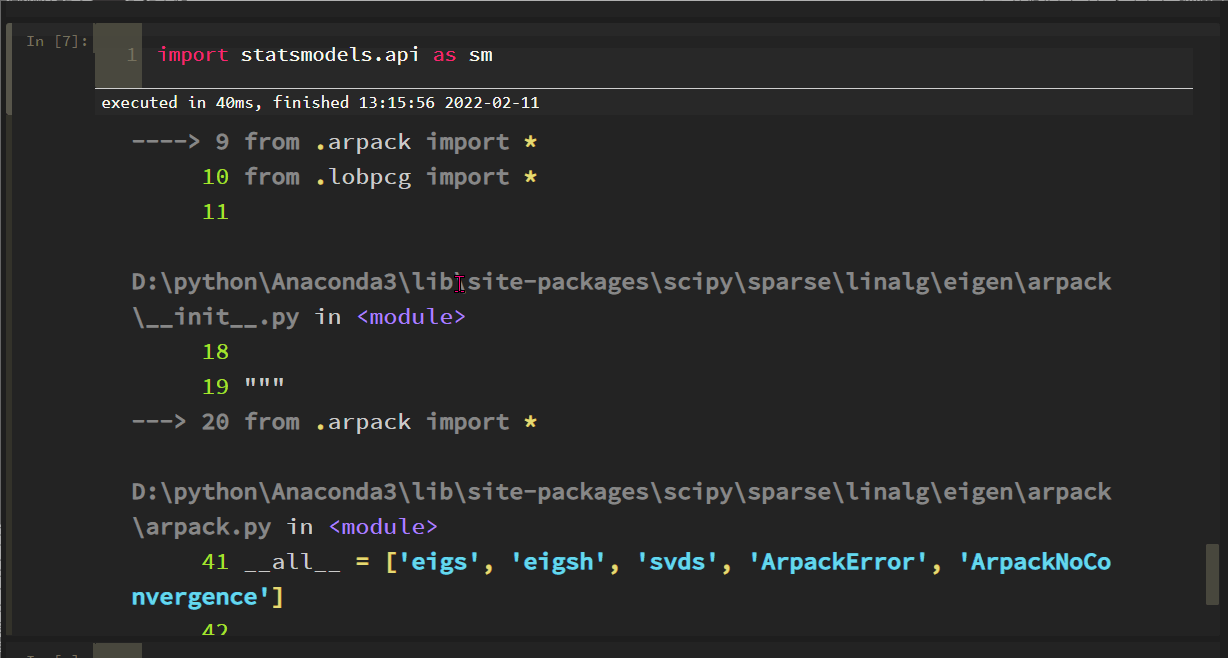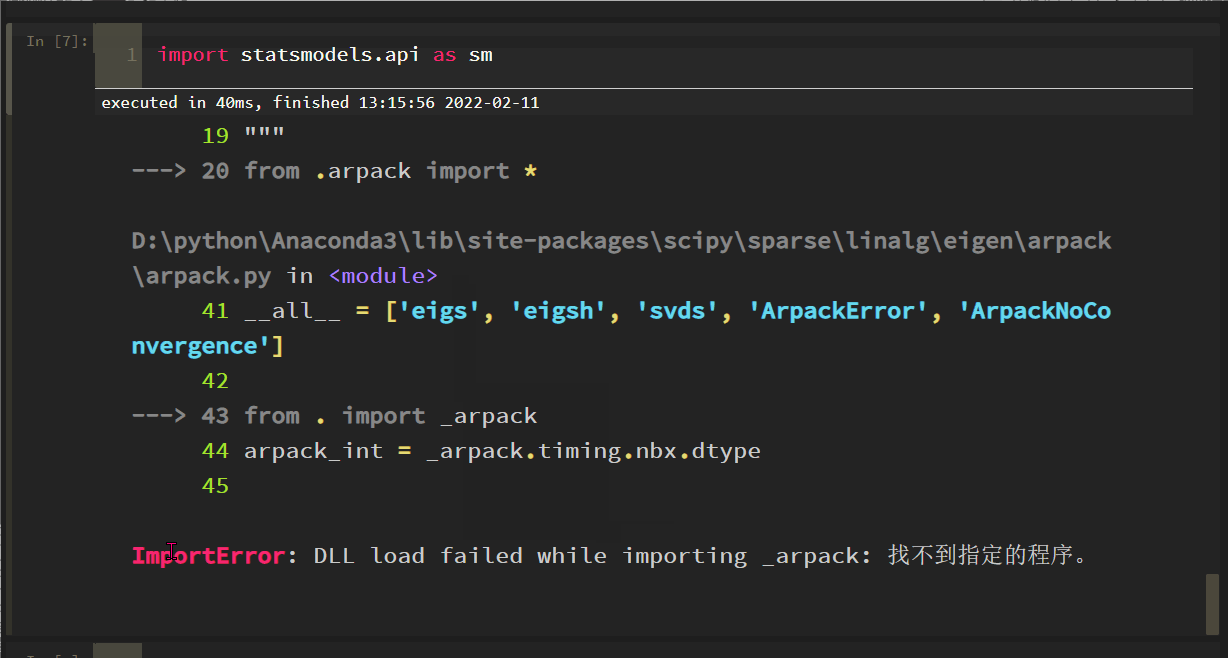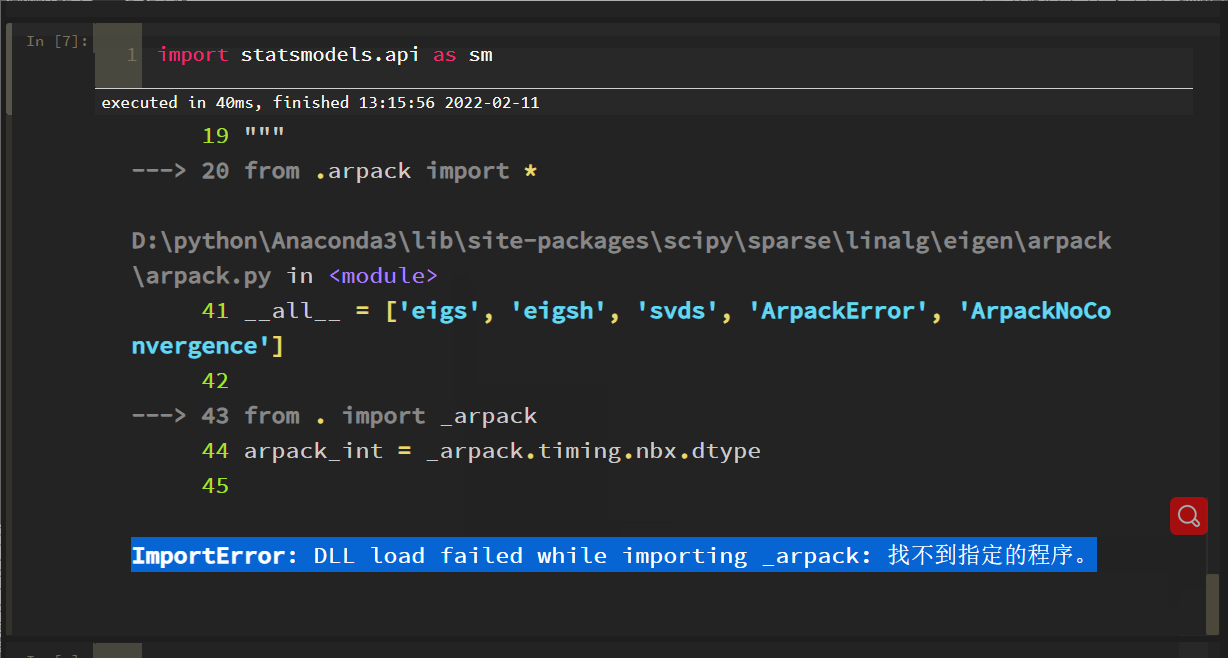
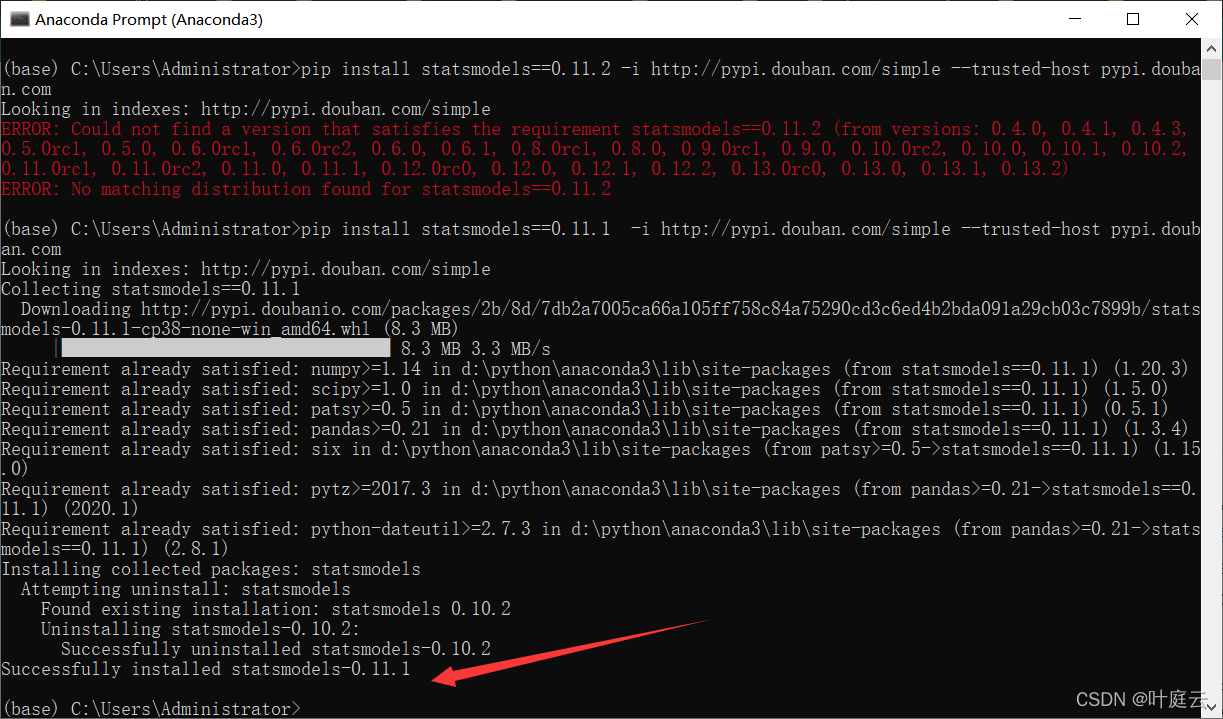
The solution process is tortuous. Roughly, the SciPy version is incompatible with some methods of statsmodels. After scipy==1.6.0, the problem is solved:
(base) C:\Users\Administrator>pip uninstall statsmodels
Found existing installation: statsmodels 0.11.1
Uninstalling statsmodels-0.11.1:
Would remove:
d:\python\anaconda3\lib\site-packages\statsmodels-0.11.1.dist-info\*
d:\python\anaconda3\lib\site-packages\statsmodels\*
Proceed (y/n)?y
Successfully uninstalled statsmodels-0.11.1
(base) C:\Users\Administrator>pip install statsmodels==0.12.0 -i http://pypi.douban.com/simple --trusted-host pypi.douban.com
Looking in indexes: http://pypi.douban.com/simple
Collecting statsmodels==0.12.0
Downloading http://pypi.doubanio.com/packages/37/22/959fdc710b6b384ad21a876130000557163978880dc50e16ba5d4ce35c18/statsmodels-0.12.0-cp38-none-win_amd64.whl (9.2 MB)
|████████████████████████████████| 9.2 MB 6.4 MB/s
Requirement already satisfied: scipy>=1.1 in d:\python\anaconda3\lib\site-packages (from statsmodels==0.12.0) (1.5.0)
Requirement already satisfied: numpy>=1.15 in d:\python\anaconda3\lib\site-packages (from statsmodels==0.12.0) (1.20.3)
Requirement already satisfied: pandas>=0.21 in d:\python\anaconda3\lib\site-packages (from statsmodels==0.12.0) (1.3.4)
Requirement already satisfied: patsy>=0.5 in d:\python\anaconda3\lib\site-packages (from statsmodels==0.12.0) (0.5.1)
Requirement already satisfied: python-dateutil>=2.7.3 in d:\python\anaconda3\lib\site-packages (from pandas>=0.21->statsmodels==0.12.0) (2.8.1)
Requirement already satisfied: pytz>=2017.3 in d:\python\anaconda3\lib\site-packages (from pandas>=0.21->statsmodels==0.12.0) (2020.1)
Requirement already satisfied: six in d:\python\anaconda3\lib\site-packages (from patsy>=0.5->statsmodels==0.12.0) (1.15.0)
Installing collected packages: statsmodels
Successfully installed statsmodels-0.12.0
(base) C:\Users\Administrator>pip show scipy
Name: scipy
Version: 1.5.0
Summary: SciPy: Scientific Library for Python
Home-page: https://www.scipy.org
Author: None
Author-email: None
License: BSD
Location: d:\python\anaconda3\lib\site-packages
Requires: numpy
Required-by: -tatsmodels, xgboost, wxgl, sweetviz, stldecompose, statsmodels, sesd, seaborn, scikit-learn, scikit-image, resampy, pyod, mplsoccer, missingno, lightgbm, librosa, imgaug, imbalanced-learn, gensim, filterpy, factor-analyzer, combo, category-encoders, catboost, bayesian-optimization, albumentations
(base) C:\Users\Administrator>pip uninstall scipy
Found existing installation: scipy 1.5.0
Uninstalling scipy-1.5.0:
Would remove:
d:\python\anaconda3\lib\site-packages\scipy-1.5.0.dist-info\*
d:\python\anaconda3\lib\site-packages\scipy\*
Proceed (y/n)?y
Successfully uninstalled scipy-1.5.0
ERROR: Exception:
Traceback (most recent call last):
File "D:\python\Anaconda3\lib\site-packages\pip\_internal\cli\base_command.py", line 188, in _main
status = self.run(options, args)
File "D:\python\Anaconda3\lib\site-packages\pip\_internal\commands\uninstall.py", line 89, in run
uninstall_pathset.commit()
File "D:\python\Anaconda3\lib\site-packages\pip\_internal\req\req_uninstall.py", line 450, in commit
self._moved_paths.commit()
File "D:\python\Anaconda3\lib\site-packages\pip\_internal\req\req_uninstall.py", line 290, in commit
save_dir.cleanup()
File "D:\python\Anaconda3\lib\site-packages\pip\_internal\utils\temp_dir.py", line 196, in cleanup
rmtree(self._path)
File "D:\python\Anaconda3\lib\site-packages\pip\_vendor\retrying.py", line 49, in wrapped_f
return Retrying(*dargs, **dkw).call(f, *args, **kw)
File "D:\python\Anaconda3\lib\site-packages\pip\_vendor\retrying.py", line 212, in call
raise attempt.get()
File "D:\python\Anaconda3\lib\site-packages\pip\_vendor\retrying.py", line 247, in get
six.reraise(self.value[0], self.value[1], self.value[2])
File "D:\python\Anaconda3\lib\site-packages\pip\_vendor\six.py", line 703, in reraise
raise value
File "D:\python\Anaconda3\lib\site-packages\pip\_vendor\retrying.py", line 200, in call
attempt = Attempt(fn(*args, **kwargs), attempt_number, False)
File "D:\python\Anaconda3\lib\site-packages\pip\_internal\utils\misc.py", line 135, in rmtree
shutil.rmtree(dir, ignore_errors=ignore_errors,
File "D:\python\Anaconda3\lib\shutil.py", line 737, in rmtree
return _rmtree_unsafe(path, onerror)
File "D:\python\Anaconda3\lib\shutil.py", line 610, in _rmtree_unsafe
_rmtree_unsafe(fullname, onerror)
File "D:\python\Anaconda3\lib\shutil.py", line 610, in _rmtree_unsafe
_rmtree_unsafe(fullname, onerror)
File "D:\python\Anaconda3\lib\shutil.py", line 615, in _rmtree_unsafe
onerror(os.unlink, fullname, sys.exc_info())
File "D:\python\Anaconda3\lib\shutil.py", line 613, in _rmtree_unsafe
os.unlink(fullname)
PermissionError: [WinError 5] Denied to Access: 'D:\\python\\Anaconda3\\Lib\\site-packages\\~cipy\\fft\\_pocketfft\\pypocketfft.cp38-win_amd64.pyd'
(base) C:\Users\Administrator>pip install scipy --user -i http://pypi.douban.com/simple --trusted-host pypi.douban.com
Looking in indexes: http://pypi.douban.com/simple
Collecting scipy
Downloading http://pypi.doubanio.com/packages/56/a3/591dbf477c35f173279afa7b9ba8e13d9c7c3d001e09aebbf6100aae33a8/scipy-1.8.0-cp38-cp38-win_amd64.whl (36.9 MB)
|████████████████████████████████| 36.9 MB 3.3 MB/s
Requirement already satisfied: numpy<1.25.0,>=1.17.3 in d:\python\anaconda3\lib\site-packages (from scipy) (1.20.3)
Installing collected packages: scipy
Successfully installed scipy-1.8.0
(base) C:\Users\Administrator>python
Python 3.8.3 (default, Jul 2 2020, 17:30:36) [MSC v.1916 64 bit (AMD64)] :: Anaconda, Inc. on win32
Type "help", "copyright", "credits" or "license" for more information.
>>> import statsmodels.api as sm
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "D:\python\Anaconda3\lib\site-packages\statsmodels\api.py", line 27, in <module>
from .tsa import api as tsa
File "D:\python\Anaconda3\lib\site-packages\statsmodels\tsa\api.py", line 31, in <module>
from .filters import api as filters
File "D:\python\Anaconda3\lib\site-packages\statsmodels\tsa\filters\api.py", line 6, in <module>
from .filtertools import miso_lfilter, convolution_filter, recursive_filter
File "D:\python\Anaconda3\lib\site-packages\statsmodels\tsa\filters\filtertools.py", line 18, in <module>
from scipy.signal.signaltools import _centered as trim_centered
ImportError: cannot import name '_centered' from 'scipy.signal.signaltools' (C:\Users\Administrator\AppData\Roaming\Python\Python38\site-packages\scipy\signal\signaltools.py)
>>> exit
Use exit() or Ctrl-Z plus Return to exit
>>> exit()
(base) C:\Users\Administrator>pip install scipy==1.6 --user -i http://pypi.douban.com/simple --trusted-host pypi.douban.co
Looking in indexes: http://pypi.douban.com/simple
WARNING: The repository located at pypi.douban.com is not a trusted or secure host and is being ignored. If this repository is available via HTTPS we recommend you use HTTPS instead, otherwise you may silence this warning and allow it anyway with '--trusted-host pypi.douban.com'.
ERROR: Could not find a version that satisfies the requirement scipy==1.6 (from versions: none)
ERROR: No matching distribution found for scipy==1.6
(base) C:\Users\Administrator>pip install scipy==1.6.0 --user -i http://pypi.douban.com/simple --trusted-host pypi.douban.com
Looking in indexes: http://pypi.douban.com/simple
Collecting scipy==1.6.0
Downloading http://pypi.doubanio.com/packages/a1/05/9b41f6a969a0d6ad36732bdd3c936434228dafaa829932be5a51574e4958/scipy-1.6.0-cp38-cp38-win_amd64.whl (32.7 MB)
|████████████████████████████████| 32.7 MB 3.2 MB/s
Requirement already satisfied: numpy>=1.16.5 in d:\python\anaconda3\lib\site-packages (from scipy==1.6.0) (1.20.3)
Installing collected packages: scipy
Attempting uninstall: scipy
Found existing installation: scipy 1.8.0
Uninstalling scipy-1.8.0:
Successfully uninstalled scipy-1.8.0
Successfully installed scipy-1.6.0
(base) C:\Users\Administrator>python
Python 3.8.3 (default, Jul 2 2020, 17:30:36) [MSC v.1916 64 bit (AMD64)] :: Anaconda, Inc. on win32
Type "help", "copyright", "credits" or "license" for more information.
>>> import statsmodels.api as sm
>>>
After the problem is solved, restart the jupyter notebook and enter it for normal use.
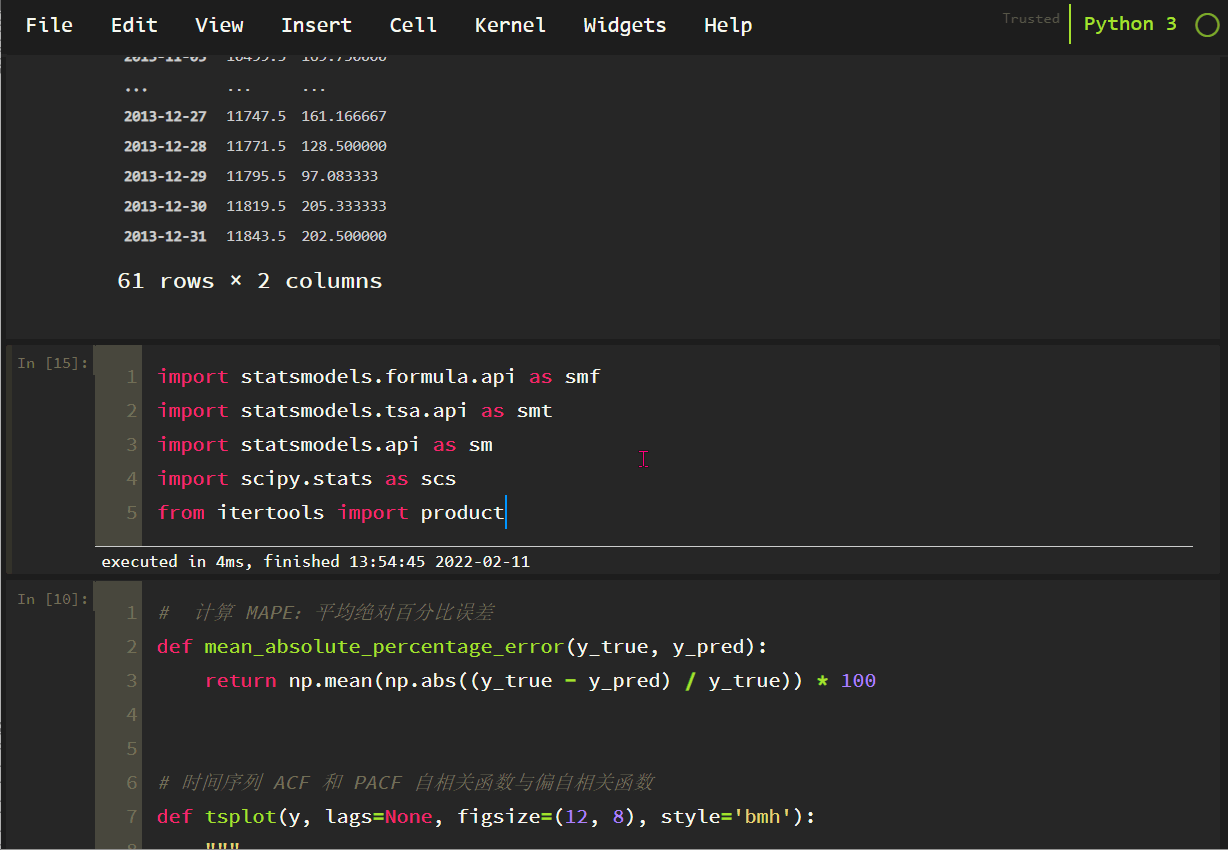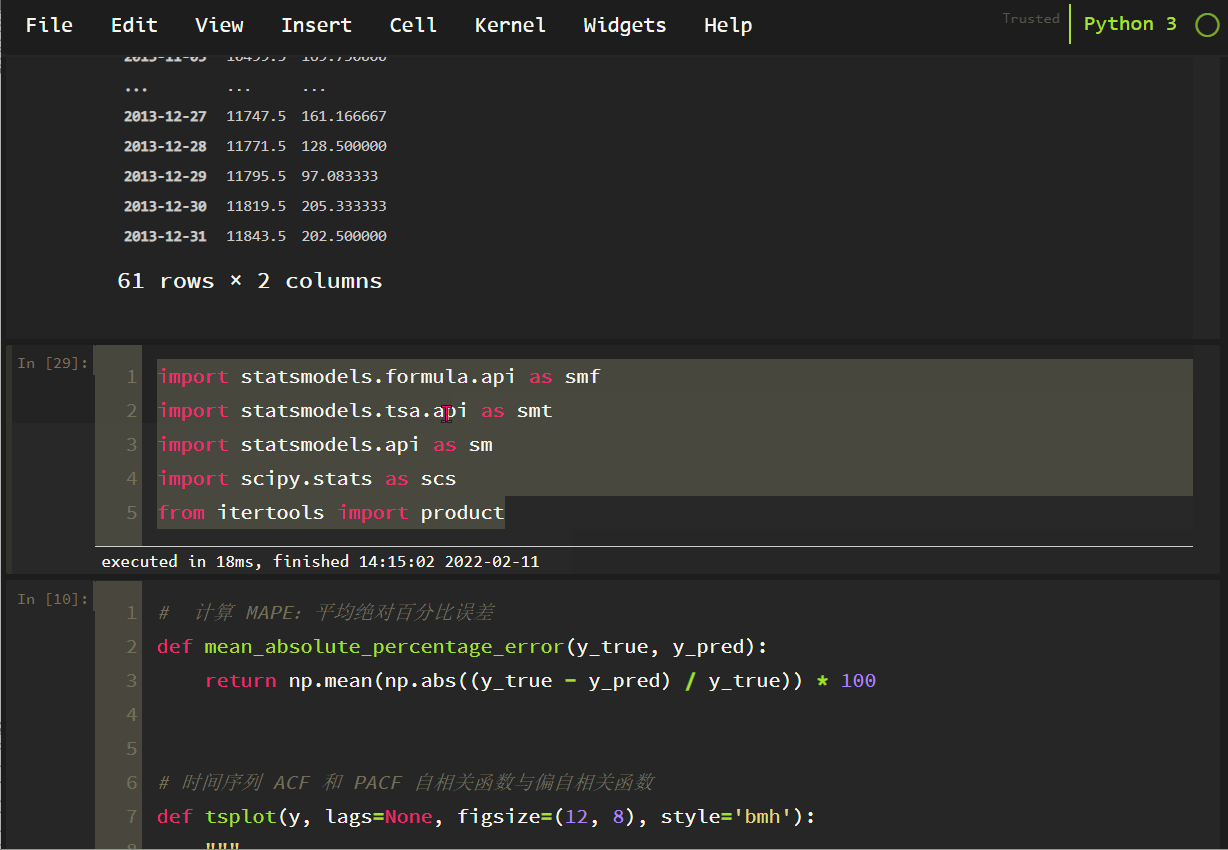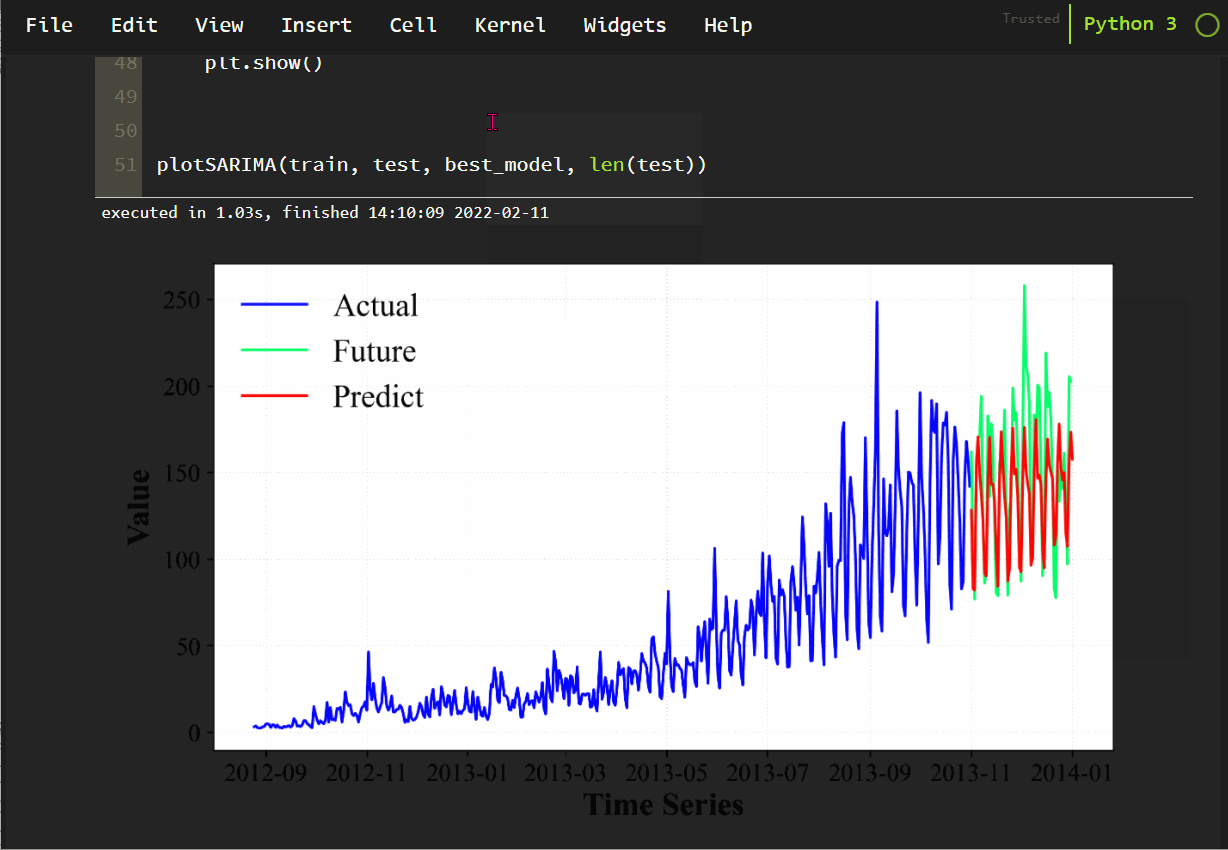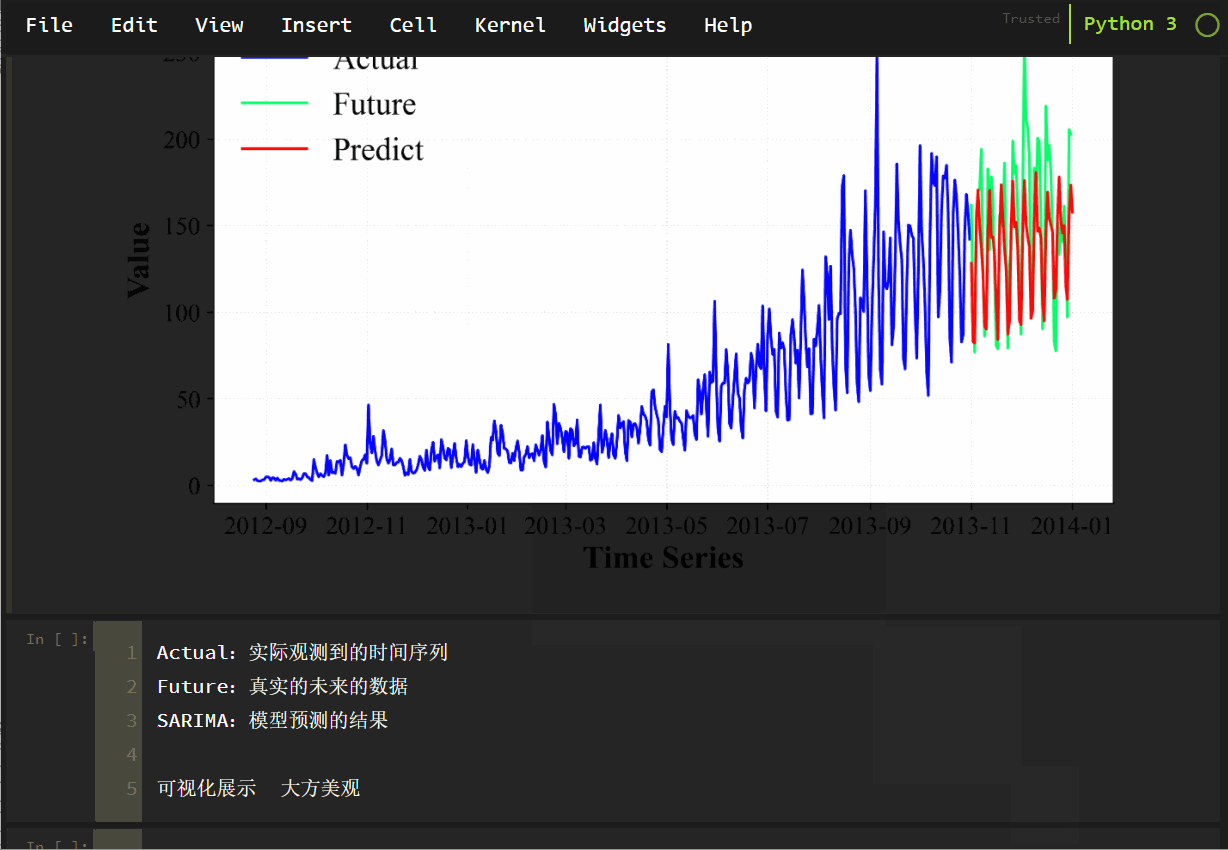

 2. Windows system & Python environment:
2. Windows system & Python environment: