try:
pd.DataFrame(output_data).to_csv(output_path, index=False)
except Exception as exp:
print("exp=",exp)
arrays must all be same length
Country_Code symbols – after removal, it seems that there is no problem
try:
pd.DataFrame(output_data).to_csv(output_path, index=False)
except Exception as exp:
print("exp=",exp)
arrays must all be same length
Country_Code symbols – after removal, it seems that there is no problem
When processing raw data, the following error occurs:
id,name,date
0,a,2020/01/01
0,b,2020/01/01
0,c,2020/01/01
0,d,2020/01/01
0,e,2020/01/01
0,f,9999/01/01
It was treated with panda :
data = pandas.read_csv(file, sep=";", encoding="ISO-8859-1", parse_dates=["date"], date_parser=lambda x: pandas.to_datetime(x, format="%d.%m.%Y"))
But the running time is wrong, which means out of bonds timestamp .
Our current approach is to skip the exception line,
The following line needs to be added
date_parser=lambda x: pd.to_datetime(x, errors="coerce")
There are three kinds of assignments for the errors parameter. The default value is’ raise ‘. An error will be reported if the parsing does not conform to the specification.
You can assign the errors parameter to “coerce” and set the time format of the error to NAT during parsing. If you don’t want to deal with the wrong time format, you can assign errors to ‘ignore’, so that the original format is the same.
errors{‘ignore’, ‘raise’, ‘coerce’}, default ‘raise’
If ‘raise’, then invalid parsing will raise an exception.If ‘coerce’, then invalid parsing will be set as NaT.If ‘ignore’, then invalid parsing will return the input.
Problem Description
After running df.groupby([‘id’])[‘click’].agg({‘click_std’: ‘std’}).reset_index(), I get nested renamer is not supported python error
Solution
In the new Pandas version, the dictionary approach of {‘click_std’:’std’} has been abandoned in favor of df.groupby([‘id ‘])[‘click’].agg(click_std=’std’).reset_index() and then run successfully.
Reference:
Restart the kernel, then execute the code again and it runs successfully!
When I want to convert a column of dates in the data frame into the date format of panda, I encountered this kind of error.
reader = pd.read_csv(f'new_files/2020-12-22-5-10.csv', usecols=['passCarTime'],dtype={'passCarTime':'string'})
pd.to_datetime(reader.passCarTime.head())
Out[98]:
0 2020-12-22 10:00:00
1 2020-12-22 10:00:00
2 2020-12-22 10:00:00
3 2020-12-22 10:00:00
4 2020-12-22 10:00:00
Name: passCarTime, dtype: datetime64[ns]
pd.to_datetime(reader.passCarTime)
Traceback (most recent call last):
File "D:\PyCharm2020\python2020\lib\site-packages\pandas\core\arrays\datetimes.py", line 2085, in objects_to_datetime64ns
values, tz_parsed = conversion.datetime_to_datetime64(data)
File "pandas\_libs\tslibs\conversion.pyx", line 350, in pandas._libs.tslibs.conversion.datetime_to_datetime64
TypeError: Unrecognized value type: <class 'str'>
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "D:\PyCharm2020\python2020\lib\site-packages\IPython\core\interactiveshell.py", line 3427, in run_code
exec(code_obj, self.user_global_ns, self.user_ns)
File "<ipython-input-99-e1b00dc18517>", line 1, in <module>
pd.to_datetime(reader.passCarTime)
File "D:\PyCharm2020\python2020\lib\site-packages\pandas\core\tools\datetimes.py", line 801, in to_datetime
cache_array = _maybe_cache(arg, format, cache, convert_listlike)
File "D:\PyCharm2020\python2020\lib\site-packages\pandas\core\tools\datetimes.py", line 178, in _maybe_cache
cache_dates = convert_listlike(unique_dates, format)
File "D:\PyCharm2020\python2020\lib\site-packages\pandas\core\tools\datetimes.py", line 465, in _convert_listlike_datetimes
result, tz_parsed = objects_to_datetime64ns(
File "D:\PyCharm2020\python2020\lib\site-packages\pandas\core\arrays\datetimes.py", line 2090, in objects_to_datetime64ns
raise e
File "D:\PyCharm2020\python2020\lib\site-packages\pandas\core\arrays\datetimes.py", line 2075, in objects_to_datetime64ns
result, tz_parsed = tslib.array_to_datetime(
File "pandas\_libs\tslib.pyx", line 364, in pandas._libs.tslib.array_to_datetime
File "pandas\_libs\tslib.pyx", line 591, in pandas._libs.tslib.array_to_datetime
File "pandas\_libs\tslib.pyx", line 726, in pandas._libs.tslib.array_to_datetime_object
File "pandas\_libs\tslib.pyx", line 717, in pandas._libs.tslib.array_to_datetime_object
File "pandas\_libs\tslibs\parsing.pyx", line 243, in pandas._libs.tslibs.parsing.parse_datetime_string
File "D:\PyCharm2020\python2020\lib\site-packages\dateutil\parser\_parser.py", line 1374, in parse
return DEFAULTPARSER.parse(timestr, **kwargs)
File "D:\PyCharm2020\python2020\lib\site-packages\dateutil\parser\_parser.py", line 649, in parse
raise ParserError("Unknown string format: %s", timestr)
dateutil.parser._parser.ParserError: Unknown string format: passCarTime
I’m not a professional, and my English is not good. I can’t understand what’s wrong. I have seen that there is no missing value in the date column of the file, and there is no date that does not conform to the format… This is very strange, welcome to leave a message, thanks in advance!
Solution
When converting, add a parameter errors ='coerce '.
reader = pd.read_csv(f'new_files/2020-12-22-5-10.csv', usecols=['passCarTime'],dtype={'passCarTime':'string'})
reader.passCarTime = pd.to_datetime(reader.passCarTime,errors='coerce')
reader.passCarTime.head()
Out[120]:
0 2020-12-22 10:00:00
1 2020-12-22 10:00:00
2 2020-12-22 10:00:00
3 2020-12-22 10:00:00
4 2020-12-22 10:00:00
Name: passCarTime, dtype: datetime64[ns]
reader.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 307707 entries, 0 to 307706
Data columns (total 1 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 passCarTime 307703 non-null datetime64[ns]
dtypes: datetime64[ns](1)
memory usage: 2.3 MB
The problem recurred
When I use pickle to reload data, all the errors are as follows:
Traceback (most recent call last):
File "segment.py", line 17, in <module>
word2id = pickle.load(pk)
File "/System/Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/pickle.py", line 1378, in load
return Unpickler(file).load()
File "/System/Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/pickle.py", line 858, in load
dispatch[key](self)
File "/System/Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/pickle.py", line 1090, in load_global
klass = self.find_class(module, name)
File "/System/Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/pickle.py", line 1124, in find_class
__import__(module)
ImportError: No module named indexes.baseThe reason for this
The same code and data run on two different machines. At first, I thought the wrong machine was missing some Python packages. But there are too many packages to install, so I can’t try them one by one. Fortunately, I use virsualenv to copy the environment from another machine to this machine directly. After running, there is no problem. But in order to find out which Python installation package is missing, I use the original compilation environment, reuse pickle to generate the original data to be loaded, and then reload it At this time, there was no error.
summary
To sum up, the reason is that the original version of panda used in the generation of pickle file is different from the current version of load pickle file. So whether it is to write code in Python or other languages, the compiling environment is very important. Once the version of a package is different, it may also lead to program errors.
Input: my_ dict = {‘i’: 1, ‘love’: 2, ‘you’: 3}
Expected output: my_ df
0
i 1
love 2
you 3If the key and value in the dictionary are one-to-one, enter my directly_ df = pd.DataFrame (my_ “Value error: if using all scalar values, you must pass an index”.
The solution is as follows:
1. Specifies the index of the dictionary when using the dataframe function
import pandas as pd
my_dict = {'i': 1, 'love': 2, 'you': 3}
my_df = pd.DataFrame(my_dict,index=[0]).T
print(my_df)
2. Convert dictionary dict to list and transfer it to dataframe
import pandas as pd
my_dict = {'i': 1, 'love': 2, 'you': 3}
my_list = [my_dict]
my_df = pd.DataFrame(my_list).T
print(my_df)
3. Use DataFrame.from_ Dict function
For specific parameters, please refer to the official website: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.from_ dict.html
import pandas as pd
my_dict = {'i': 1, 'love': 2, 'you': 3}
my_df = pd.DataFrame.from_dict(my_dict, orient='index')
print(my_df)Output results
0
i 1
love 2
you 3A very convenient way to use pandas is through indexes such as LOC, iloc and IX. here is a record:
df.loc [condition, new column] = assign initial value
If the new column name is an existing column name, it will be changed on the original data column
import pandas as pd
import numpy as np
data = pd.DataFrame ( np.random.randint (0,100,40).reshape(10,4),columns=list(‘abcd’))
print(data)
data.loc [data. D & gt; = 50, ‘greater than 50′] =’Yes’
Print (data)
By using LOC to index, judge in the index, and then assign value to the new column according to the result of judgment. This is a very convenient and basic operation. Of course, I don’t remember it clearly recently, so I’ll record it here.
The reason is that XLRD was recently updated to version 2.0.1 and only supports.xls files. So Pandas. Read_Excel (‘ xxx.xlsx ‘) will return an error.
You can install an older version of XLRD and run it in CMD:
PIP uninstall XLRD
PIP install XLRD ==1.2.0
You can also open the.xlsx file with OpenPyXL instead of XLRD:
Df = pandas read_excel (‘ data. XLSX, engine = ‘openpyxl)
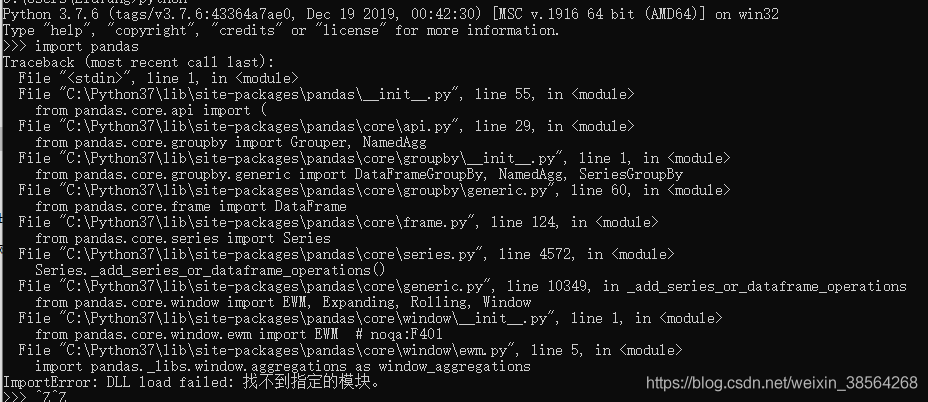
pip uninstall pandasThen specify the PIP to download version 1.0.1 of Pandas
pip install pandas==1.0.1
Convention:
import pandas as pd
import numpy as npReIndex reindexes
Reindex () is an important method of the Pandas object, which creates a new object with a new index.
I. Re-index Series objects
se1=pd.Series([1,7,3,9],index=['d','c','a','f'])
se1Code results:
d 1
c 7
a 3
f 9
dtype: int64
Calling reindex will reorder the missing values and fill them with NaN.
se2=se1.reindex(['a','b','c','d','e','f'])
se2Code results:
a 3.0
b NaN
c 7.0
d 1.0
e NaN
f 9.0
dtype: float64
When passing in method= “” select interpolation processing mode when reindexing:
Method = ‘ffill’ or ‘pad forward filling
Method = ‘bfill’ or ‘backfill’
se3=pd.Series(['blue','red','black'],index=[0,2,4])
se4=se3.reindex(range(6),method='ffill')
se4Code results:
0 blue
1 blue
2 red
3 red
4 black
5 black
dtype: object
Second, reindex the DataFrame object
For a DataFrame object, reIndex can modify the row index and column index.
df1=pd.DataFrame(np.arange(9).reshape(3,3),index=['a','c','d'],columns=['one','two','four'])
df1Code results:
| one | two | four | |
|---|---|---|---|
| a | 0 | 1 | 2 |
| c | 3 | 4 | 5 |
| d | 6 | 7 | 8 |
Reordering the row index by default
Passing in only one sequence does not rearrange the index of the sequence
df1.reindex(['a','b','c','d'])Code results:
The
| one | two | four | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a 0.0 | 1.0 | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| b | NaN | NaN | NaN | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4.0 | 5.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| d 6.0 | 7.0 | 8.0
Code results:
|