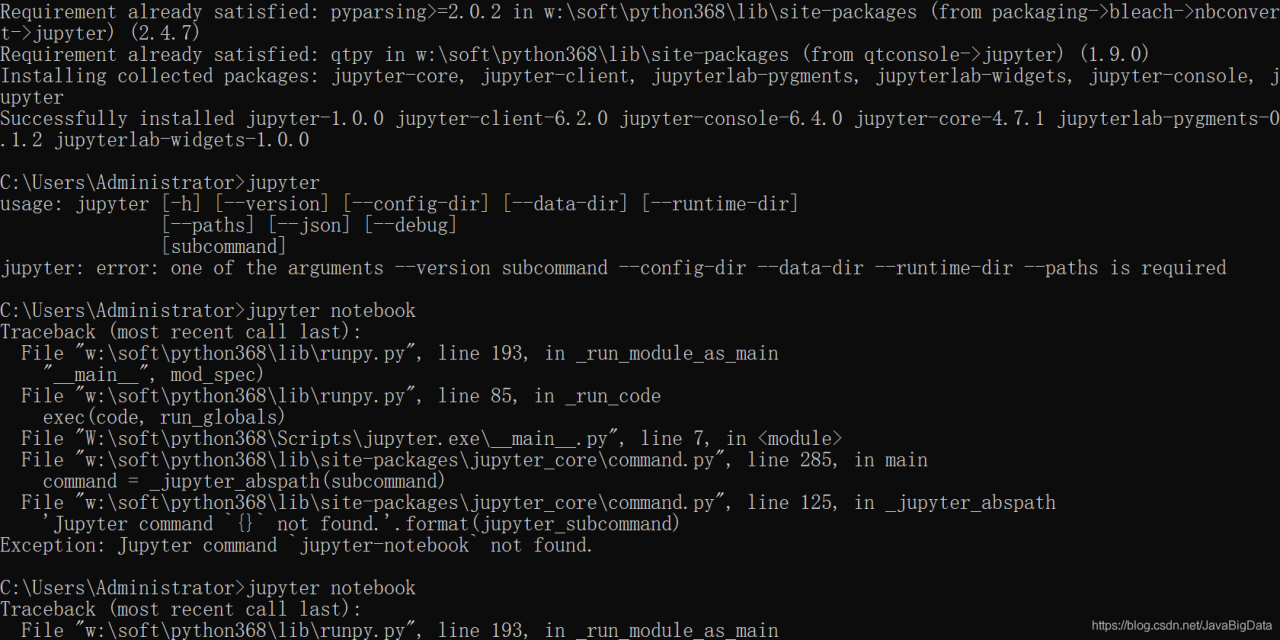
Before I installed the keras environment on my computer, it ran successfully, but there was an error when I installed it again on the Ubuntu server, and the error as shown in the title appeared when I ran the code. By looking at the file in the error keras, we found a problem:
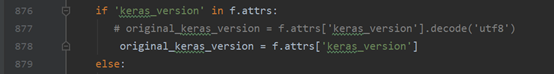
 . This file is in “… Envs/tensorf/lib/site packages/keras/engine/saving. Py”. Figure 1 does not need to encode the data, but figure 2 does. According to other blogs, encode encodes data into machine language, while decode decodes machine language into high-level language. The error is that I installed keras as saving. Py in the second version of figure (strange that I installed it on my computer and server). You just need to delete the “decode (‘utf8 ‘)” in the wrong lines. It seems that 3-4 places need to be deleted
. This file is in “… Envs/tensorf/lib/site packages/keras/engine/saving. Py”. Figure 1 does not need to encode the data, but figure 2 does. According to other blogs, encode encodes data into machine language, while decode decodes machine language into high-level language. The error is that I installed keras as saving. Py in the second version of figure (strange that I installed it on my computer and server). You just need to delete the “decode (‘utf8 ‘)” in the wrong lines. It seems that 3-4 places need to be deleted
attachment: the revised saving. Py code is as follows:
"""Model saving utilities.
"""
from __future__ import print_function
from __future__ import absolute_import
from __future__ import division
import numpy as np
import os
import json
import yaml
import warnings
from six.moves import zip
from .. import backend as K
from .. import optimizers
from ..utils.io_utils import ask_to_proceed_with_overwrite
from ..utils import conv_utils
try:
import h5py
HDF5_OBJECT_HEADER_LIMIT = 64512
except ImportError:
h5py = None
def save_model(model, filepath, overwrite=True, include_optimizer=True):
"""Save a model to a HDF5 file.
Note: Please also see
[How can I install HDF5 or h5py to save my models in Keras?](
/getting-started/faq/
#how-can-i-install-HDF5-or-h5py-to-save-my-models-in-Keras)
in the FAQ for instructions on how to install `h5py`.
The saved model contains:
- the model's configuration (topology)
- the model's weights
- the model's optimizer's state (if any)
Thus the saved model can be reinstantiated in
the exact same state, without any of the code
used for model definition or training.
# Arguments
model: Keras model instance to be saved.
filepath: one of the following:
- string, path where to save the model, or
- h5py.File object where to save the model
overwrite: Whether we should overwrite any existing
model at the target location, or instead
ask the user with a manual prompt.
include_optimizer: If True, save optimizer's state together.
# Raises
ImportError: if h5py is not available.
"""
if h5py is None:
raise ImportError('`save_model` requires h5py.')
def get_json_type(obj):
"""Serialize any object to a JSON-serializable structure.
# Arguments
obj: the object to serialize
# Returns
JSON-serializable structure representing `obj`.
# Raises
TypeError: if `obj` cannot be serialized.
"""
# if obj is a serializable Keras class instance
# e.g. optimizer, layer
if hasattr(obj, 'get_config'):
return {'class_name': obj.__class__.__name__,
'config': obj.get_config()}
# if obj is any numpy type
if type(obj).__module__ == np.__name__:
if isinstance(obj, np.ndarray):
return {'type': type(obj),
'value': obj.tolist()}
else:
return obj.item()
# misc functions (e.g. loss function)
if callable(obj):
return obj.__name__
# if obj is a python 'type'
if type(obj).__name__ == type.__name__:
return obj.__name__
raise TypeError('Not JSON Serializable:', obj)
from .. import __version__ as keras_version
if not isinstance(filepath, h5py.File):
# If file exists and should not be overwritten.
if not overwrite and os.path.isfile(filepath):
proceed = ask_to_proceed_with_overwrite(filepath)
if not proceed:
return
f = h5py.File(filepath, mode='w')
opened_new_file = True
else:
f = filepath
opened_new_file = False
try:
f.attrs['keras_version'] = str(keras_version).encode('utf8')
f.attrs['backend'] = K.backend().encode('utf8')
f.attrs['model_config'] = json.dumps({
'class_name': model.__class__.__name__,
'config': model.get_config()
}, default=get_json_type).encode('utf8')
model_weights_group = f.create_group('model_weights')
model_layers = model.layers
save_weights_to_hdf5_group(model_weights_group, model_layers)
if include_optimizer and model.optimizer:
if isinstance(model.optimizer, optimizers.TFOptimizer):
warnings.warn(
'TensorFlow optimizers do not '
'make it possible to access '
'optimizer attributes or optimizer state '
'after instantiation. '
'As a result, we cannot save the optimizer '
'as part of the model save file.'
'You will have to compile your model again '
'after loading it. '
'Prefer using a Keras optimizer instead '
'(see keras.io/optimizers).')
else:
f.attrs['training_config'] = json.dumps({
'optimizer_config': {
'class_name': model.optimizer.__class__.__name__,
'config': model.optimizer.get_config()
},
'loss': model.loss,
'metrics': model.metrics,
'sample_weight_mode': model.sample_weight_mode,
'loss_weights': model.loss_weights,
}, default=get_json_type).encode('utf8')
# Save optimizer weights.
symbolic_weights = getattr(model.optimizer, 'weights')
if symbolic_weights:
optimizer_weights_group = f.create_group(
'optimizer_weights')
weight_values = K.batch_get_value(symbolic_weights)
weight_names = []
for i, (w, val) in enumerate(zip(symbolic_weights,
weight_values)):
# Default values of symbolic_weights is /variable
# for Theano and CNTK
if K.backend() == 'theano' or K.backend() == 'cntk':
if hasattr(w, 'name'):
if w.name.split('/')[-1] == 'variable':
name = str(w.name) + '_' + str(i)
else:
name = str(w.name)
else:
name = 'param_' + str(i)
else:
if hasattr(w, 'name') and w.name:
name = str(w.name)
else:
name = 'param_' + str(i)
weight_names.append(name.encode('utf8'))
optimizer_weights_group.attrs[
'weight_names'] = weight_names
for name, val in zip(weight_names, weight_values):
param_dset = optimizer_weights_group.create_dataset(
name,
val.shape,
dtype=val.dtype)
if not val.shape:
# scalar
param_dset[()] = val
else:
param_dset[:] = val
f.flush()
finally:
if opened_new_file:
f.close()
def load_model(filepath, custom_objects=None, compile=True):
"""Loads a model saved via `save_model`.
# Arguments
filepath: one of the following:
- string, path to the saved model, or
- h5py.File object from which to load the model
custom_objects: Optional dictionary mapping names
(strings) to custom classes or functions to be
considered during deserialization.
compile: Boolean, whether to compile the model
after loading.
# Returns
A Keras model instance. If an optimizer was found
as part of the saved model, the model is already
compiled. Otherwise, the model is uncompiled and
a warning will be displayed. When `compile` is set
to False, the compilation is omitted without any
warning.
# Raises
ImportError: if h5py is not available.
ValueError: In case of an invalid savefile.
"""
if h5py is None:
raise ImportError('`load_model` requires h5py.')
if not custom_objects:
custom_objects = {}
def convert_custom_objects(obj):
"""Handles custom object lookup.
# Arguments
obj: object, dict, or list.
# Returns
The same structure, where occurrences
of a custom object name have been replaced
with the custom object.
"""
if isinstance(obj, list):
deserialized = []
for value in obj:
deserialized.append(convert_custom_objects(value))
return deserialized
if isinstance(obj, dict):
deserialized = {}
for key, value in obj.items():
deserialized[key] = convert_custom_objects(value)
return deserialized
if obj in custom_objects:
return custom_objects[obj]
return obj
opened_new_file = not isinstance(filepath, h5py.File)
if opened_new_file:
f = h5py.File(filepath, mode='r')
else:
f = filepath
model = None
try:
# instantiate model
model_config = f.attrs.get('model_config')
if model_config is None:
raise ValueError('No model found in config file.')
# model_config = json.loads(model_config.decode('utf-8'))
model_config = json.loads(model_config)
model = model_from_config(model_config, custom_objects=custom_objects)
# set weights
load_weights_from_hdf5_group(f['model_weights'], model.layers)
if compile:
# instantiate optimizer
training_config = f.attrs.get('training_config')
if training_config is None:
warnings.warn('No training configuration found in save file: '
'the model was *not* compiled. '
'Compile it manually.')
return model
# training_config = json.loads(training_config.decode('utf-8'))
training_config = json.loads(training_config)
optimizer_config = training_config['optimizer_config']
optimizer = optimizers.deserialize(optimizer_config,
custom_objects=custom_objects)
# Recover loss functions and metrics.
loss = convert_custom_objects(training_config['loss'])
metrics = convert_custom_objects(training_config['metrics'])
sample_weight_mode = training_config['sample_weight_mode']
loss_weights = training_config['loss_weights']
# Compile model.
model.compile(optimizer=optimizer,
loss=loss,
metrics=metrics,
loss_weights=loss_weights,
sample_weight_mode=sample_weight_mode)
# Set optimizer weights.
if 'optimizer_weights' in f:
# Build train function (to get weight updates).
model._make_train_function()
optimizer_weights_group = f['optimizer_weights']
optimizer_weight_names = [
n.decode('utf8') for n in
optimizer_weights_group.attrs['weight_names']]
optimizer_weight_values = [optimizer_weights_group[n] for n in
optimizer_weight_names]
try:
model.optimizer.set_weights(optimizer_weight_values)
except ValueError:
warnings.warn('Error in loading the saved optimizer '
'state. As a result, your model is '
'starting with a freshly initialized '
'optimizer.')
finally:
if opened_new_file:
f.close()
return model
def model_from_config(config, custom_objects=None):
"""Instantiates a Keras model from its config.
# Arguments
config: Configuration dictionary.
custom_objects: Optional dictionary mapping names
(strings) to custom classes or functions to be
considered during deserialization.
# Returns
A Keras model instance (uncompiled).
# Raises
TypeError: if `config` is not a dictionary.
"""
if isinstance(config, list):
raise TypeError('`model_from_config` expects a dictionary, '
'not a list. Maybe you meant to use '
'`Sequential.from_config(config)`?')
from ..layers import deserialize
return deserialize(config, custom_objects=custom_objects)
def model_from_yaml(yaml_string, custom_objects=None):
"""Parses a yaml model configuration file and returns a model instance.
# Arguments
yaml_string: YAML string encoding a model configuration.
custom_objects: Optional dictionary mapping names
(strings) to custom classes or functions to be
considered during deserialization.
# Returns
A Keras model instance (uncompiled).
"""
config = yaml.load(yaml_string)
from ..layers import deserialize
return deserialize(config, custom_objects=custom_objects)
def model_from_json(json_string, custom_objects=None):
"""Parses a JSON model configuration file and returns a model instance.
# Arguments
json_string: JSON string encoding a model configuration.
custom_objects: Optional dictionary mapping names
(strings) to custom classes or functions to be
considered during deserialization.
# Returns
A Keras model instance (uncompiled).
"""
config = json.loads(json_string)
from ..layers import deserialize
return deserialize(config, custom_objects=custom_objects)
def save_attributes_to_hdf5_group(group, name, data):
"""Saves attributes (data) of the specified name into the HDF5 group.
This method deals with an inherent problem of HDF5 file which is not
able to store data larger than HDF5_OBJECT_HEADER_LIMIT bytes.
# Arguments
group: A pointer to a HDF5 group.
name: A name of the attributes to save.
data: Attributes data to store.
"""
# Check that no item in `data` is larger than `HDF5_OBJECT_HEADER_LIMIT`
# because in that case even chunking the array would not make the saving
# possible.
bad_attributes = [x for x in data if len(x) > HDF5_OBJECT_HEADER_LIMIT]
# Expecting this to never be true.
if len(bad_attributes) > 0:
raise RuntimeError('The following attributes cannot be saved to HDF5 '
'file because they are larger than %d bytes: %s'
% (HDF5_OBJECT_HEADER_LIMIT,
', '.join([x for x in bad_attributes])))
data_npy = np.asarray(data)
num_chunks = 1
chunked_data = np.array_split(data_npy, num_chunks)
# This will never loop forever thanks to the test above.
while any(map(lambda x: x.nbytes > HDF5_OBJECT_HEADER_LIMIT, chunked_data)):
num_chunks += 1
chunked_data = np.array_split(data_npy, num_chunks)
if num_chunks > 1:
for chunk_id, chunk_data in enumerate(chunked_data):
group.attrs['%s%d' % (name, chunk_id)] = chunk_data
else:
group.attrs[name] = data
def load_attributes_from_hdf5_group(group, name):
"""Loads attributes of the specified name from the HDF5 group.
This method deals with an inherent problem
of HDF5 file which is not able to store
data larger than HDF5_OBJECT_HEADER_LIMIT bytes.
# Arguments
group: A pointer to a HDF5 group.
name: A name of the attributes to load.
# Returns
data: Attributes data.
"""
if name in group.attrs:
data = [n.decode('utf8') for n in group.attrs[name]]
else:
data = []
chunk_id = 0
while ('%s%d' % (name, chunk_id)) in group.attrs:
data.extend([n.decode('utf8')
for n in group.attrs['%s%d' % (name, chunk_id)]])
chunk_id += 1
return data
def save_weights_to_hdf5_group(f, layers):
from .. import __version__ as keras_version
save_attributes_to_hdf5_group(
f, 'layer_names', [layer.name.encode('utf8') for layer in layers])
f.attrs['backend'] = K.backend().encode('utf8')
f.attrs['keras_version'] = str(keras_version).encode('utf8')
for layer in layers:
g = f.create_group(layer.name)
symbolic_weights = layer.weights
weight_values = K.batch_get_value(symbolic_weights)
weight_names = []
for i, (w, val) in enumerate(zip(symbolic_weights, weight_values)):
if hasattr(w, 'name') and w.name:
name = str(w.name)
else:
name = 'param_' + str(i)
weight_names.append(name.encode('utf8'))
save_attributes_to_hdf5_group(g, 'weight_names', weight_names)
for name, val in zip(weight_names, weight_values):
param_dset = g.create_dataset(name, val.shape,
dtype=val.dtype)
if not val.shape:
# scalar
param_dset[()] = val
else:
param_dset[:] = val
def preprocess_weights_for_loading(layer, weights,
original_keras_version=None,
original_backend=None,
reshape=False):
"""Converts layers weights from Keras 1 format to Keras 2 and also weights of CuDNN layers in Keras 2.
# Arguments
layer: Layer instance.
weights: List of weights values (Numpy arrays).
original_keras_version: Keras version for the weights, as a string.
original_backend: Keras backend the weights were trained with,
as a string.
reshape: Reshape weights to fit the layer when the correct number
of values are present but the shape does not match.
# Returns
A list of weights values (Numpy arrays).
"""
def convert_nested_bidirectional(weights):
"""Converts layers nested in `Bidirectional` wrapper by `preprocess_weights_for_loading()`.
# Arguments
weights: List of weights values (Numpy arrays).
# Returns
A list of weights values (Numpy arrays).
"""
num_weights_per_layer = len(weights) // 2
forward_weights = preprocess_weights_for_loading(layer.forward_layer,
weights[:num_weights_per_layer],
original_keras_version,
original_backend)
backward_weights = preprocess_weights_for_loading(layer.backward_layer,
weights[num_weights_per_layer:],
original_keras_version,
original_backend)
return forward_weights + backward_weights
def convert_nested_time_distributed(weights):
"""Converts layers nested in `TimeDistributed` wrapper by `preprocess_weights_for_loading()`.
# Arguments
weights: List of weights values (Numpy arrays).
# Returns
A list of weights values (Numpy arrays).
"""
return preprocess_weights_for_loading(
layer.layer, weights, original_keras_version, original_backend)
def convert_nested_model(weights):
"""Converts layers nested in `Model` or `Sequential` by `preprocess_weights_for_loading()`.
# Arguments
weights: List of weights values (Numpy arrays).
# Returns
A list of weights values (Numpy arrays).
"""
new_weights = []
# trainable weights
for sublayer in layer.layers:
num_weights = len(sublayer.trainable_weights)
if num_weights > 0:
new_weights.extend(preprocess_weights_for_loading(
layer=sublayer,
weights=weights[:num_weights],
original_keras_version=original_keras_version,
original_backend=original_backend))
weights = weights[num_weights:]
# non-trainable weights
for sublayer in layer.layers:
num_weights = len([l for l in sublayer.weights
if l not in sublayer.trainable_weights])
if num_weights > 0:
new_weights.extend(preprocess_weights_for_loading(
layer=sublayer,
weights=weights[:num_weights],
original_keras_version=original_keras_version,
original_backend=original_backend))
weights = weights[num_weights:]
return new_weights
# Convert layers nested in Bidirectional/TimeDistributed/Model/Sequential.
# Both transformation should be ran for both Keras 1->2 conversion
# and for conversion of CuDNN layers.
if layer.__class__.__name__ == 'Bidirectional':
weights = convert_nested_bidirectional(weights)
if layer.__class__.__name__ == 'TimeDistributed':
weights = convert_nested_time_distributed(weights)
elif layer.__class__.__name__ in ['Model', 'Sequential']:
weights = convert_nested_model(weights)
if original_keras_version == '1':
if layer.__class__.__name__ == 'TimeDistributed':
weights = preprocess_weights_for_loading(layer.layer,
weights,
original_keras_version,
original_backend)
if layer.__class__.__name__ == 'Conv1D':
shape = weights[0].shape
# Handle Keras 1.1 format
if shape[:2] != (layer.kernel_size[0], 1) or shape[3] != layer.filters:
# Legacy shape:
# (filters, input_dim, filter_length, 1)
assert shape[0] == layer.filters and shape[2:] == (layer.kernel_size[0], 1)
weights[0] = np.transpose(weights[0], (2, 3, 1, 0))
weights[0] = weights[0][:, 0, :, :]
if layer.__class__.__name__ == 'Conv2D':
if layer.data_format == 'channels_first':
# old: (filters, stack_size, kernel_rows, kernel_cols)
# new: (kernel_rows, kernel_cols, stack_size, filters)
weights[0] = np.transpose(weights[0], (2, 3, 1, 0))
if layer.__class__.__name__ == 'Conv2DTranspose':
if layer.data_format == 'channels_last':
# old: (kernel_rows, kernel_cols, stack_size, filters)
# new: (kernel_rows, kernel_cols, filters, stack_size)
weights[0] = np.transpose(weights[0], (0, 1, 3, 2))
if layer.data_format == 'channels_first':
# old: (filters, stack_size, kernel_rows, kernel_cols)
# new: (kernel_rows, kernel_cols, filters, stack_size)
weights[0] = np.transpose(weights[0], (2, 3, 0, 1))
if layer.__class__.__name__ == 'Conv3D':
if layer.data_format == 'channels_first':
# old: (filters, stack_size, ...)
# new: (..., stack_size, filters)
weights[0] = np.transpose(weights[0], (2, 3, 4, 1, 0))
if layer.__class__.__name__ == 'GRU':
if len(weights) == 9:
kernel = np.concatenate([weights[0],
weights[3],
weights[6]], axis=-1)
recurrent_kernel = np.concatenate([weights[1],
weights[4],
weights[7]], axis=-1)
bias = np.concatenate([weights[2],
weights[5],
weights[8]], axis=-1)
weights = [kernel, recurrent_kernel, bias]
if layer.__class__.__name__ == 'LSTM':
if len(weights) == 12:
# old: i, c, f, o
# new: i, f, c, o
kernel = np.concatenate([weights[0],
weights[6],
weights[3],
weights[9]], axis=-1)
recurrent_kernel = np.concatenate([weights[1],
weights[7],
weights[4],
weights[10]], axis=-1)
bias = np.concatenate([weights[2],
weights[8],
weights[5],
weights[11]], axis=-1)
weights = [kernel, recurrent_kernel, bias]
if layer.__class__.__name__ == 'ConvLSTM2D':
if len(weights) == 12:
kernel = np.concatenate([weights[0],
weights[6],
weights[3],
weights[9]], axis=-1)
recurrent_kernel = np.concatenate([weights[1],
weights[7],
weights[4],
weights[10]], axis=-1)
bias = np.concatenate([weights[2],
weights[8],
weights[5],
weights[11]], axis=-1)
if layer.data_format == 'channels_first':
# old: (filters, stack_size, kernel_rows, kernel_cols)
# new: (kernel_rows, kernel_cols, stack_size, filters)
kernel = np.transpose(kernel, (2, 3, 1, 0))
recurrent_kernel = np.transpose(recurrent_kernel,
(2, 3, 1, 0))
weights = [kernel, recurrent_kernel, bias]
conv_layers = ['Conv1D',
'Conv2D',
'Conv3D',
'Conv2DTranspose',
'ConvLSTM2D']
if layer.__class__.__name__ in conv_layers:
layer_weights_shape = K.int_shape(layer.weights[0])
if _need_convert_kernel(original_backend):
weights[0] = conv_utils.convert_kernel(weights[0])
if layer.__class__.__name__ == 'ConvLSTM2D':
weights[1] = conv_utils.convert_kernel(weights[1])
if reshape and layer_weights_shape != weights[0].shape:
if weights[0].size != np.prod(layer_weights_shape):
raise ValueError('Weights must be of equal size to ' +
'apply a reshape operation. ' +
'Layer ' + layer.name +
'\'s weights have shape ' +
str(layer_weights_shape) + ' and size ' +
str(np.prod(layer_weights_shape)) + '. ' +
'The weights for loading have shape ' +
str(weights[0].shape) + ' and size ' +
str(weights[0].size) + '. ')
weights[0] = np.reshape(weights[0], layer_weights_shape)
elif layer_weights_shape != weights[0].shape:
weights[0] = np.transpose(weights[0], (3, 2, 0, 1))
if layer.__class__.__name__ == 'ConvLSTM2D':
weights[1] = np.transpose(weights[1], (3, 2, 0, 1))
# convert CuDNN layers
weights = _convert_rnn_weights(layer, weights)
return weights
def _convert_rnn_weights(layer, weights):
"""Converts weights for RNN layers between native and CuDNN format.
Input kernels for each gate are transposed and converted between Fortran
and C layout, recurrent kernels are transposed. For LSTM biases are summed/
split in half, for GRU biases are reshaped.
Weights can be converted in both directions between `LSTM` and`CuDNNSLTM`
and between `CuDNNGRU` and `GRU(reset_after=True)`. Default `GRU` is not
compatible with `CuDNNGRU`.
For missing biases in `LSTM`/`GRU` (`use_bias=False`),
no conversion is made.
# Arguments
layer: Target layer instance.
weights: List of source weights values (input kernels, recurrent
kernels, [biases]) (Numpy arrays).
# Returns
A list of converted weights values (Numpy arrays).
# Raises
ValueError: for incompatible GRU layer/weights or incompatible biases
"""
def transform_kernels(kernels, func, n_gates):
"""Transforms kernel for each gate separately using given function.
# Arguments
kernels: Stacked array of kernels for individual gates.
func: Function applied to kernel of each gate.
n_gates: Number of gates (4 for LSTM, 3 for GRU).
# Returns
Stacked array of transformed kernels.
"""
return np.hstack([func(k) for k in np.hsplit(kernels, n_gates)])
def transpose_input(from_cudnn):
"""Makes a function that transforms input kernels from/to CuDNN format.
It keeps the shape, but changes between the layout (Fortran/C). Eg.:
```
Keras CuDNN
[[0, 1, 2], <---> [[0, 2, 4],
[3, 4, 5]] [1, 3, 5]]
```
It can be passed to `transform_kernels()`.
# Arguments
from_cudnn: `True` if source weights are in CuDNN format, `False`
if they're in plain Keras format.
# Returns
Function that converts input kernel to the other format.
"""
order = 'F' if from_cudnn else 'C'
def transform(kernel):
return kernel.T.reshape(kernel.shape, order=order)
return transform
target_class = layer.__class__.__name__
# convert the weights between CuDNNLSTM and LSTM
if target_class in ['LSTM', 'CuDNNLSTM'] and len(weights) == 3:
# determine if we're loading a CuDNNLSTM layer
# from the number of bias weights:
# CuDNNLSTM has (units * 8) weights; while LSTM has (units * 4)
# if there's no bias weight in the file, skip this conversion
units = weights[1].shape[0]
bias_shape = weights[2].shape
n_gates = 4
if bias_shape == (2 * units * n_gates,):
source = 'CuDNNLSTM'
elif bias_shape == (units * n_gates,):
source = 'LSTM'
else:
raise ValueError('Invalid bias shape: ' + str(bias_shape))
def convert_weights(weights, from_cudnn=True):
# transpose (and reshape) input and recurrent kernels
kernels = transform_kernels(weights[0],
transpose_input(from_cudnn),
n_gates)
recurrent_kernels = transform_kernels(weights[1], lambda k: k.T, n_gates)
if from_cudnn:
# merge input and recurrent biases into a single set
biases = np.sum(np.split(weights[2], 2, axis=0), axis=0)
else:
# Split single set of biases evenly to two sets. The way of
# splitting doesn't matter as long as the two sets sum is kept.
biases = np.tile(0.5 * weights[2], 2)
return [kernels, recurrent_kernels, biases]
if source != target_class:
weights = convert_weights(weights, from_cudnn=source == 'CuDNNLSTM')
# convert the weights between CuDNNGRU and GRU(reset_after=True)
if target_class in ['GRU', 'CuDNNGRU'] and len(weights) == 3:
# We can determine the source of the weights from the shape of the bias.
# If there is no bias we skip the conversion since CuDNNGRU always has biases.
units = weights[1].shape[0]
bias_shape = weights[2].shape
n_gates = 3
def convert_weights(weights, from_cudnn=True):
kernels = transform_kernels(weights[0], transpose_input(from_cudnn), n_gates)
recurrent_kernels = transform_kernels(weights[1], lambda k: k.T, n_gates)
biases = np.array(weights[2]).reshape((2, -1) if from_cudnn else -1)
return [kernels, recurrent_kernels, biases]
if bias_shape == (2 * units * n_gates,):
source = 'CuDNNGRU'
elif bias_shape == (2, units * n_gates):
source = 'GRU(reset_after=True)'
elif bias_shape == (units * n_gates,):
source = 'GRU(reset_after=False)'
else:
raise ValueError('Invalid bias shape: ' + str(bias_shape))
if target_class == 'CuDNNGRU':
target = 'CuDNNGRU'
elif layer.reset_after:
target = 'GRU(reset_after=True)'
else:
target = 'GRU(reset_after=False)'
# only convert between different types
if source != target:
types = (source, target)
if 'GRU(reset_after=False)' in types:
raise ValueError('%s is not compatible with %s' % types)
if source == 'CuDNNGRU':
weights = convert_weights(weights, from_cudnn=True)
elif source == 'GRU(reset_after=True)':
weights = convert_weights(weights, from_cudnn=False)
return weights
def _need_convert_kernel(original_backend):
"""Checks if conversion on kernel matrices is required during weight loading.
The convolution operation is implemented differently in different backends.
While TH implements convolution, TF and CNTK implement the correlation operation.
So the channel axis needs to be flipped when we're loading TF weights onto a TH model,
or vice verca. However, there's no conversion required between TF and CNTK.
# Arguments
original_backend: Keras backend the weights were trained with, as a string.
# Returns
`True` if conversion on kernel matrices is required, otherwise `False`.
"""
if original_backend is None:
# backend information not available
return False
uses_correlation = {'tensorflow': True,
'theano': False,
'cntk': True}
if original_backend not in uses_correlation:
# By default, do not convert the kernels if the original backend is unknown
return False
if K.backend() in uses_correlation:
current_uses_correlation = uses_correlation[K.backend()]
else:
# Assume unknown backends use correlation
current_uses_correlation = True
return uses_correlation[original_backend] != current_uses_correlation
def load_weights_from_hdf5_group(f, layers, reshape=False):
"""Implements topological (order-based) weight loading.
# Arguments
f: A pointer to a HDF5 group.
layers: a list of target layers.
reshape: Reshape weights to fit the layer when the correct number
of values are present but the shape does not match.
# Raises
ValueError: in case of mismatch between provided layers
and weights file.
"""
if 'keras_version' in f.attrs:
# original_keras_version = f.attrs['keras_version'].decode('utf8')
original_keras_version = f.attrs['keras_version']
else:
original_keras_version = '1'
if 'backend' in f.attrs:
# original_backend = f.attrs['backend'].decode('utf8')
original_backend = f.attrs['backend']
else:
original_backend = None
filtered_layers = []
for layer in layers:
weights = layer.weights
if weights:
filtered_layers.append(layer)
layer_names = load_attributes_from_hdf5_group(f, 'layer_names')
filtered_layer_names = []
for name in layer_names:
g = f[name]
weight_names = load_attributes_from_hdf5_group(g, 'weight_names')
if weight_names:
filtered_layer_names.append(name)
layer_names = filtered_layer_names
if len(layer_names) != len(filtered_layers):
raise ValueError('You are trying to load a weight file '
'containing ' + str(len(layer_names)) +
' layers into a model with ' +
str(len(filtered_layers)) + ' layers.')
# We batch weight value assignments in a single backend call
# which provides a speedup in TensorFlow.
weight_value_tuples = []
for k, name in enumerate(layer_names):
g = f[name]
weight_names = load_attributes_from_hdf5_group(g, 'weight_names')
weight_values = [np.asarray(g[weight_name]) for weight_name in weight_names]
layer = filtered_layers[k]
symbolic_weights = layer.weights
weight_values = preprocess_weights_for_loading(layer,
weight_values,
original_keras_version,
original_backend,
reshape=reshape)
if len(weight_values) != len(symbolic_weights):
raise ValueError('Layer #' + str(k) +
' (named "' + layer.name +
'" in the current model) was found to '
'correspond to layer ' + name +
' in the save file. '
'However the new layer ' + layer.name +
' expects ' + str(len(symbolic_weights)) +
' weights, but the saved weights have ' +
str(len(weight_values)) +
' elements.')
weight_value_tuples += zip(symbolic_weights, weight_values)
K.batch_set_value(weight_value_tuples)
def load_weights_from_hdf5_group_by_name(f, layers, skip_mismatch=False,
reshape=False):
"""Implements name-based weight loading.
(instead of topological weight loading).
Layers that have no matching name are skipped.
# Arguments
f: A pointer to a HDF5 group.
layers: A list of target layers.
skip_mismatch: Boolean, whether to skip loading of layers
where there is a mismatch in the number of weights,
or a mismatch in the shape of the weights.
reshape: Reshape weights to fit the layer when the correct number
of values are present but the shape does not match.
# Raises
ValueError: in case of mismatch between provided layers
and weights file and skip_mismatch=False.
"""
if 'keras_version' in f.attrs:
original_keras_version = f.attrs['keras_version'].decode('utf8')
else:
original_keras_version = '1'
if 'backend' in f.attrs:
original_backend = f.attrs['backend'].decode('utf8')
else:
original_backend = None
# New file format.
layer_names = load_attributes_from_hdf5_group(f, 'layer_names')
# Reverse index of layer name to list of layers with name.
index = {}
for layer in layers:
if layer.name:
index.setdefault(layer.name, []).append(layer)
# We batch weight value assignments in a single backend call
# which provides a speedup in TensorFlow.
weight_value_tuples = []
for k, name in enumerate(layer_names):
g = f[name]
weight_names = load_attributes_from_hdf5_group(g, 'weight_names')
weight_values = [np.asarray(g[weight_name]) for weight_name in weight_names]
for layer in index.get(name, []):
symbolic_weights = layer.weights
weight_values = preprocess_weights_for_loading(
layer,
weight_values,
original_keras_version,
original_backend,
reshape=reshape)
if len(weight_values) != len(symbolic_weights):
if skip_mismatch:
warnings.warn('Skipping loading of weights for layer {}'.format(layer.name) +
' due to mismatch in number of weights' +
' ({} vs {}).'.format(len(symbolic_weights), len(weight_values)))
continue
else:
raise ValueError('Layer #' + str(k) +
' (named "' + layer.name +
'") expects ' +
str(len(symbolic_weights)) +
' weight(s), but the saved weights' +
' have ' + str(len(weight_values)) +
' element(s).')
# Set values.
for i in range(len(weight_values)):
if K.int_shape(symbolic_weights[i]) != weight_values[i].shape:
if skip_mismatch:
warnings.warn('Skipping loading of weights for layer {}'.format(layer.name) +
' due to mismatch in shape' +
' ({} vs {}).'.format(
symbolic_weights[i].shape,
weight_values[i].shape))
continue
else:
raise ValueError('Layer #' + str(k) +
' (named "' + layer.name +
'"), weight ' +
str(symbolic_weights[i]) +
' has shape {}'.format(K.int_shape(symbolic_weights[i])) +
', but the saved weight has shape ' +
str(weight_values[i].shape) + '.')
else:
weight_value_tuples.append((symbolic_weights[i],
weight_values[i]))
K.batch_set_value(weight_value_tuples)

 . According to the above figure, I find that my computer’s CUDA version is 10.0, so I will report an error at runtime. At this time, there are two solutions.
. According to the above figure, I find that my computer’s CUDA version is 10.0, so I will report an error at runtime. At this time, there are two solutions.
 . This file is in “… Envs/tensorf/lib/site packages/keras/engine/saving. Py”. Figure 1 does not need to encode the data, but figure 2 does. According to other blogs, encode encodes data into machine language, while decode decodes machine language into high-level language. The error is that I installed keras as saving. Py in the second version of figure (strange that I installed it on my computer and server). You just need to delete the “decode (‘utf8 ‘)” in the wrong lines. It seems that 3-4 places need to be deleted
. This file is in “… Envs/tensorf/lib/site packages/keras/engine/saving. Py”. Figure 1 does not need to encode the data, but figure 2 does. According to other blogs, encode encodes data into machine language, while decode decodes machine language into high-level language. The error is that I installed keras as saving. Py in the second version of figure (strange that I installed it on my computer and server). You just need to delete the “decode (‘utf8 ‘)” in the wrong lines. It seems that 3-4 places need to be deleted